Abstract
Context
Polyunsaturated fatty acids (PUFA) are important during pregnancy for fetal development and child health outcomes. The fatty acid desaturase (FADS) genes also influence PUFA status, with the FADS genes controlling how much product (eg, arachidonic acid, eicosapentaenoic acid, and docosahexaenoic acid) is metabolized from the precursor molecules linoleic acid and α-linolenic acid.
Objective
The current review discusses the influence of FADS genotype on PUFA status of pregnant women, breast milk, and children, and also how FADS may influence child health outcomes.
Data sources
The Ovid Medline, Scopus, Embase, Cochrane Library, CINAHL Plus, PubMed and Web of Science databases were searched from their inception to September 2018.
Data extraction
Eligible studies reported FADS genotype and blood concentrations of PUFA during pregnancy, in childhood, breast milk concentrations of PUFA or child health outcomes.
Data analysis
In pregnant and lactating women, minor allele carriers have higher concentrations of linoleic acid and α-linolenic acid, and lower concentrations of arachidonic acid, in blood and breast milk, respectively. In children, FADS genotype influences PUFA status in the same manner and may impact child outcomes such as cognition and allergies; however, the direction of effects for the evidence to date is not consistent.
Conclusion
Further studies are needed to further investigate associations between FADS and outcomes, as well as the diet-gene interaction.
Keywords: fatty acid desaturase, FADS, polyunsaturated fatty acids, PUFA, pregnancy, child outcomes
INTRODUCTION
Polyunsaturated fatty acids (PUFA) have many important roles in physiological processes in the body, including cell membrane function, response to inflammation,1 and fetal development.1,2 As the developing fetus relies solely on maternal supply for nutrition, maternal status of PUFA during pregnancy is of particular importance. Long chain (LC) PUFA include arachidonic acid (AA) and docosahexaenoic acid (DHA) of the n-6 and n-3 pathways, respectively. DHA is required for brain and retinal development, and AA is essential for brain growth.3–5 DHA and AA are found in high concentrations in the gray matter of the brain and membranes of the retina.6 AA has a role in cell signaling and division,7 and accumulates in the brain during development, particularly from the third trimester to 2 years of age.8 Animal studies have shown that AA is involved in maintenance and functioning of the hippocampus9 and also has a protective role against oxidative stress.10
In the latter stages of pregnancy, levels of DHA and AA in the developing fetal brain rise, while levels of the precursors polyunsaturated fatty acids (PUFA) α-linolenic acid (ALA) and linolenic acid (LA) remain stable.3 A recent systematic review highlighted the effects of n-3 LCPUFA supplementation during pregnancy, which was found to result in a lower incidence of preterm birth.11 LCPUFA supplementation during pregnancy has also been linked to a reduction in childhood allergic diseases,12,13 an increase in childhood fat-free mass,14 protection against increased blood pressure in children who are overweight or obese,15 and cognitive and visual development16; however, the evidence has not shown a conclusive benefit. DHA and AA are also required postnatally for child development. Both DHA and AA have been found to be associated with cognitive and visual function outcomes in children.17 Supplementation of infant formula with DHA+AA has been associated with higher scores on the Mental Development Index and the Bayley Scales of Infant Development, 2nd edition.18
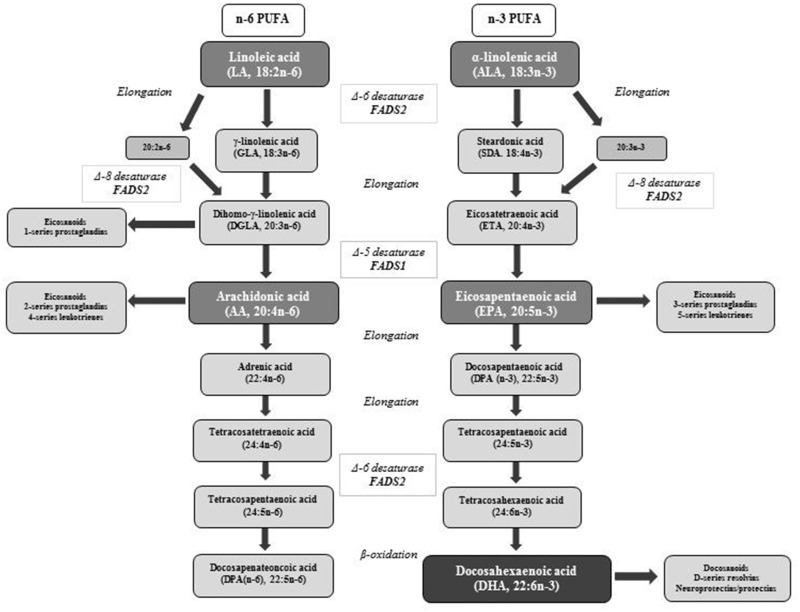
Increasing evidence suggests that genetic variation in fatty acid desaturase (FADS) genes is associated with biological PUFA concentrations. The FADS1 and FADS2 genes are found on chromosome 11q12–q13.1 in head-to-head orientation, and together with FADS3, they form the FADS gene cluster.19 These FADS genes are involved in the PUFA metabolic pathway (Figure 1). The rate-limiting desaturase enzymes involved in this pathway include Δ-5 desaturase (D5D) and Δ-6 desaturase (D6D), which are encoded by the FADS1 and FADS2 genes, respectively.20,21 D5D and D6D enzyme activity can be estimated as fatty acid product:precursor ratios.22 The n-6 and n-3 families compete for the limited desaturase23,24 and elongase enzymes.25,26 The desaturase enzymes have a preference for n-3 PUFA; however, owing to the high dietary intake of LA, there is higher desaturation to n-6 PUFA.24
Figure 1.
Metabolic synthesis of polyunsaturated fatty acids (amended from Yeates et al [2015]20). Abbreviations: FADS1, fatty acid desaturase 1 gene; FADS2, fatty acid desaturase 2 gene; n-3 PUFA, polyunsaturated fatty acid of the n-3 pathway; n-6 PUFA, polyunsaturated fatty acid of the n-6 pathway.
Genetic variation in FADS genotype is commonly observed with single-nucleotide polymorphisms (SNPs), resulting in modifications to biological status and ability to synthesize LCPUFA. Results from 7 observational studies indicate that pregnant women who are carriers of the minor allele of various FADS SNPs have higher blood concentrations of the precursors LA and ALA and lower concentrations of LCPUFA, in particular AA.20,27–33 Studies also indicate that minor allele carriers have lower desaturase activity, as indicated by reduced product:precursor ratios.27,30 This reduction in enzyme activity may be due to a decrease in functional enzymes apparent with the presence of the variant allele. There are, however, other possible mechanisms of action of FADS SNPs on PUFA status that operate via changes in FADS gene expression. Such mechanisms include altered promoter or enhancer region of the FADS gene, transcript degradation,34,35 and low expression of protein.36
FADS genotype variation has also been shown to have an effect on breast-milk PUFA concentrations in lactating women. A total of 7 observational studies have investigated the relationship between maternal FADS genotype and breast-milk PUFA composition. Minor allele carriage in a range of FADS SNPs was associated with lower AA breast milk concentrations.32,33,37–40 Lower concentrations of eicosapentaenoic acid (EPA)32,33,38 and DHA32,33 have been reported in some, but not all, studies in this area.
Child PUFA status is also influenced by FADS genotype, with a total of 12 relevant studies in this area. PUFA status of children – as measured in cord blood according to genotype – has been investigated in 3 studies,41–43 and in children aged 2–13 years in 4 studies.41,44–46 Children who were minor allele carriers for FADS SNPs generally had decreased circulating AA concentrations. Associations of child FADS genotype and child DHA status have not been as consistent, with minor allele carriers of some SNPs having increased DHA concentrations, while other FADS SNPs are associated with decreased DHA concentrations.47 Other studies have reported no association between DHA status and FADS genotype in children aged 8 years and over.46,48 Maternal FADS genotype has also been found to influence child PUFA status. Maternal genotype has been associated with increased cord blood LA concentrations, as well as decreased AA and DHA,43 AA to DGLA (dihomo-γ-linolenic acid) ratios, and EPA to ALA ratios.42
In addition to influencing PUFA status, FADS genotype variation and associations with clinical endpoints have also been examined. One of the most commonly investigated outcomes in relation to child FADS genotype is child cognition. No direct association between child FADS genotype and cognitive outcomes has been reported.40,44,48–50 The influence of child FADS on allergy development has also been investigated, similarly with no conclusive relationship observed.41,45,51 Other child health outcomes that are known to be associated with PUFA status, such as inflammatory marker status,38 asthma,52 and lipid profile,53,54 have also been studied but the research in these areas to date is limited.
To date, there has been no systematic review assessing the effects of maternal and child FADS genotype on PUFA status in both mother and infant, and how this might impact on child outcomes. The current review has discussed the existing evidence in relation to the influence of genetic variation in FADS genotype on PUFA status of pregnant women, breast milk, and children, and on child outcomes including, but not limited to, cognition, allergy development, and asthma. The need for further studies in relation to FADS genotype and PUFA status and child outcomes is highlighted.
OBJECTIVES
The current review aimed to evaluate the current scientific literature on FADS genotype and its influence on PUFA status in pregnancy, during lactation, and in childhood. The influence of FADS genotype on child outcomes was also reviewed.
METHODS
Search methods for identification of studies
The Ovid Medline, Embase, Scopus, Cochrane Library, CINAHL Plus, PubMed, and Web of Science databases were searched from their inception until September 2018. Search terms included truncated and MESH (medical subject heading) terms where appropriate. The search strategy consisted of several elements: FADS, PUFA, and either pregnancy, lactation and breast milk; children; or child outcomes (see Appendix S1 in the Supporting Information online). Searches were modified as required for each database. A secondary search of reference lists for studies viewed as eligible for inclusion was also completed whereby potentially relevant articles were identified.
Eligibility criteria
Articles were viewed as eligible if they reported FADS genotype and at least one of the following outcomes: blood concentrations of PUFA during pregnancy, lactation, or childhood; breast milk concentrations of PUFA; and child health outcomes (including cognition, birth outcomes, allergy development, or other relevant outcomes). Studies completed in pregnant women, lactating women, or children were eligible for inclusion. Observational studies were included if they examined FADS genotype and PUFA status in the previously mentioned population groups or if they examined FADS genotype and child outcomes. A PICOS strategy (participants, intervention, comparison, outcomes, and study design) was developed to define inclusion and exclusion criteria for intervention studies (Table 1). For logistical reasons, only articles published in the English language were viewed as eligible.
Table 1.
PICOS criteria for inclusion and exclusion of studies
| Parameter | Inclusion criteria | Exclusion criteria |
|---|---|---|
| Population | Pregnant or lactating women, women of childbearing age, children | Adult males only |
| Intervention | Fish oil supplements, intervention with fish, fatty acid supplements | No consideration of fish oil supplements, intervention with fish, fatty acid supplements |
| Comparison | Different FADS genotype groups | No comparison of FADS genotype groups |
| Outcomes | PUFA status of women during pregnancy; PUFA status of lactating mothers; PUFA status of breast-milk composition; child PUFA status; child development outcomes such as cognition, ADHD, allergies, and immunity | Outcomes other than PUFA status, or child health outcomes |
| Study design | Observational studies, RCTs, non-RCTs | Animal studies; in vitro studies; drug studies; chemical interaction studies; laboratory studies; food technology studies; cell culture studies; method development articles; research policy/policy-making articles; proof-of-concept articles; letters; editorials; commentaries; studies not published in the English language |
Abbreviations: ADHD, attention deficit hyperactivity disorder; FADS, fatty acid desaturase; PUFA, polyunsaturated fatty acid; RCT, randomized controlled trial.
Study selection and data extraction
All database search results were exported to RefWorks and duplicate records were removed. Titles were screened for eligibility using the above predefined eligibility screening criteria. Following title and abstract screening, full texts for each article were obtained. Full-text articles were subsequently assessed for eligibility. Reference lists of relevant studies were also searched for potentially eligible papers; these were then screened as described previously. Relevant data was extracted from individual studies, including country, study participants, study type, sample size, FADS SNPs, and study findings.
Quality assessment
As a means of assessing the quality of eligible studies, the Newcastle-Ottawa scale was used to evaluate observational studies55 – specifically, the areas of selection, comparability, exposure, and outcome. Studies were scored according to a predefined star scoring system, where a higher star rating indicated a better quality study, with a maximum possible score of 9 stars. The Cochrane Collaboration tool for assessing risk of bias was used to assess the quality of intervention studies.56 This risk of bias tool assessed the areas of sequence generation, allocation concealment, blinding, incomplete data, and selective reporting. Studies were judged to have a low, high, or unclear risk of bias in each of these areas.
RESULTS
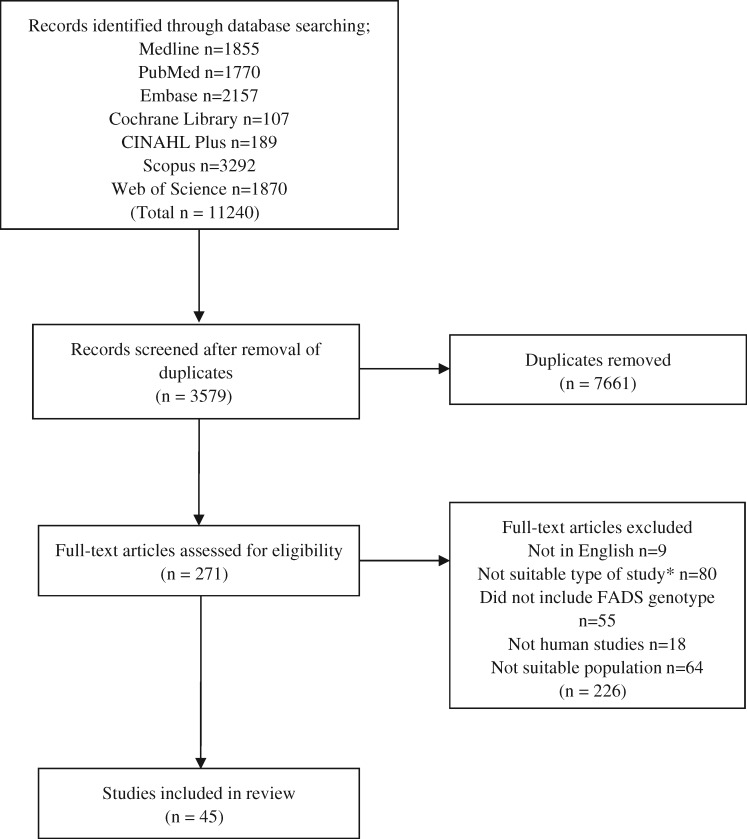
A total of 11 240 papers were identified from the database searches (Figure 2). Of these, 7661 were excluded as they were duplicates, leaving 3579 references. Following title and abstract screening, 271 full-text articles were assessed for eligibility. A further 226 references were excluded following full-text screening: 9 were not written in English and 80 were not a suitable study type (reviews/conference proceedings/abstract only/meta-analysis/book chapter), or the full text was not available. A further 55 did not include FADS genotype, 18 were not human studies, and 64 were studies completed in a population outside the scope of this review (eg, studies in men, or women who were not pregnant). Following these exclusions, 45 articles were deemed suitable for inclusion in the current review.
Figure 2.
Flow diagram of the literature search process. *Studies considered to be not suitable included reviews, conference proceedings, abstracts only, meta-analyses, and book chapters, or the full text was not available. Abbreviation: FADS, fatty acid desaturase.
Following quality assessment for observational studies using the Newcastle-Ottawa Scale, the majority of observational studies were determined to be of good quality, with scores for selection generally being high (see Tables S1 and S2 in the Supporting Information online). Quality scores were generally lower in the area of comparability owing to studies not controlling for important confounders. The majority of studies scored highly in the outcome or exposure category. For the included intervention studies, 2 of the 3 studies included in this review had an unclear risk of selection bias (see Table S3 in the Supporting Information online). Overall, there was a low risk of performance and detection bias. Attrition bias was varied in each intervention study included in this review: one had a high risk of bias, one an unclear risk of bias, and one a low risk of bias. All studies were judged to have an unclear risk of reporting bias.
DISCUSSION
FADS genotype and PUFA status during pregnancy
During pregnancy, the developing fetus relies solely on maternal supply for nutrients, which are transferred from mother to fetus via placental transfer. Eight articles investigated the influence of FADS genotype on PUFA status during pregnancy (Table 2). All authors reported increased concentrations of the n-6 precursor molecule LA and n-3 ALA in minor allele carriers of various FADS SNPs. Minor allele carriers for rs174545, rs174537, rs174546, and rs174553 of the FADS1 gene had lower AA:DGLA ratio, indicative of decreased functionality of the desaturase enzymes.27 Similarly, others have shown a negative association between minor allele carriers of a range of SNPs and product:substrate ratios (Table 2), further suggestive of decreased enzyme activity in the presence of FADS genotype SNPs. An intervention study in pregnant women investigated the association between FADS1 SNP rs174533 and FADS2 SNP rs174575 and AA and DHA status following DHA supplementation.57 At baseline, minor allele carriers for rs174533 had lower AA and DHA status; however, following supplementation with DHA the minor allele homozygotes had increased DHA status from baseline. This finding suggests that genotype influences the effect of dietary intake on PUFA status. As these data were derived from a randomized controlled trial, they provide an insight into the influence of supplements on PUFA status, rather than being observational data looking at one point in time. Pregnant women who are minor allele carriers of FADS SNPs may therefore benefit from increased dietary intake of preformed LCPUFA for improved circulating LCPUFA concentrations important for fetal development. Of these studies, 6 reported PUFA status as determined from the phospholipid fraction of either plasma or red blood cells, while 2 reported total PUFA concentrations from serum or plasma. During pregnancy, maternal PUFAs are transferred from the mother to the fetus via the placenta. These PUFAs arise from maternal triglycerides and free fatty acids rather than phospholipids.58–60 Consideration of the blood fraction being used for analysis should be carefully considered to ensure the appropriate fraction is chosen. Given the importance of PUFA for fetal development, optimal maternal status is crucial and future dietary intervention studies are needed to further investigate the influence of dietary intake on PUFA status according to genotype.
Table 2.
Associations between maternal FADS genotype and maternal PUFA status during pregnancy
| Reference (country) | Participants, blood fraction for PUFA analysis (study type, n) | FADS gene, SNP | Findings |
|---|---|---|---|
| de la Garza Puentes et al (2017)27 (Spain) | Pregnant women, plasma phospholipids; 24 wk gestation (cohort study, n = 180) | FADS1 rs174537, rs174545, rs174546, rs174553, rs174547, rs174548, rs174561; FADS2 rs1535, rs174575, rs174583, rs99780, rs174602 | Minor allele carriers of rs174545, rs174537, rs174546, and rs174553 were negatively associated with AA:DGLA ratio. Minor allele carriers of rs174545, rs174546, and rs174553 had significantly lower AA concentrations than major allele carriers. |
| Koletzko et al (2011)30 (England) | Pregnant women, RBC phospholipids; 4–44 wk gestation (longitudinal study, n = 4457) | FADS1 rs174548, rs174556, rs174561; intergenic rs3834458, rs968567; FADS2 rs174570, rs174574, rs2727271, rs174576, rs174578, rs174579, rs174602, rs498793, rs526126; intergenic rs174448, rs174449; FADS3 rs174455 | Positive association between minor allele carriers and precursor FAs and a negative association with LCPUFA products. Minor allele carriers of rs174548, rs174556, rs174561, rs3834458, rs968567, rs174570, rs174574, rs2727271, rs174576, rs174579, rs174602, rs526126, rs174448, rs174449, and rs174455 were negatively associated with product:substrate ratios for n-6 and n-3 pathways. A significant association was found between minor allele carriers and lower RBC DHA. |
| Xie and Innis (2008)33 (Canada) | Pregnant women, plasma phospholipids, RBC ethanolamine phosphoglycerides; 16 and 36 wk gestation (observational study, n = 69) | FADS1 intergenic rs174553; FADS2 rs99780, rs174575, rs174583; haplotypes for FADS1/FADS2 | Minor allele homozygotes of rs174553, rs99780, and rs174583 had lower plasma phospholipid and erythrocyte concentrations of AA but higher concentrations of LA. Minor allele carriers also had decreased n-6 and n-3 product:precursor ratios at both 16 and 36 weeks’ gestation. |
| Xie and Innis (2009)31 (Canada) | Pregnant women, plasma phospholipids, RBC ethanolamine phosphoglycerides; 16 wk gestation (observational study, n = 69) | FADS1 intergenic rs174553; FADS1 rs99780; FADS2 rs174575, rs174583; haplotypes for FADS1/FADS2 | Minor allele carriers of rs174553, rs99780, and rs174583 had lower AA and higher concentrations of LA than major allele carriers. Minor allele haplotypes were associated with higher LA and lower AA in plasma and RBC ethanolamine phosphoglycerides. |
| Moltó-Puigmartí et al (2010)32 (the Netherlands) | Pregnant women, plasma phospholipids; 36 wk gestation (observational study, n = 309) | FADS1 rs174561; FADS2 rs174575; intergenic rs3834458; haplotypes for FADS1/FADS2 | Minor allele homozygotes had significantly higher LA and lower AA. For the n-3 fatty acids, higher ALA and lower EPA and DPA concentrations were observed; however, these associations did not meet the same level of significance as the n-6 fatty acids. Haplotype analysis results were similar to results for single SNPs, with homozygous minor alleles associated with lower concentrations of product molecules such as AA, and higher concentrations of precursor LA, compared with major allele homozygotes. |
| Yeates et al (2015)20 (Seychelles) | Pregnant women and their offspring, total serum; 28 wk gestation (longitudinal study, n = 1622) | FADS1 rs174537, rs174561; FADS1–FADS2 rs3834458; FADS2 rs174575 | rs3834458 was significantly associated with AA and LA:AA ratio where an increasing number of variant alleles resulted in decreased AA concentrations. rs174575 minor allele carriers had a higher LA:AA ratio than major allele carriers, and lower AA concentrations. No significant associations were seen between genotype and EPA, DHA, or ratio of ALA:EPA. |
| Gonzalez-Casanova et al (2016)28 (Mexico) | Pregnant women and their offspring, total plasma; 18–22 wk gestation (RCT – baseline data only for PUFA, n = 654) | FADS1 rs174556; FADS2 rs174602, rs498793; FADS3 rs174455 | Maternal rs174455, rs174556, and rs174602 were positively associated with maternal AA concentrations. rs174556 was positively associated with DHA, whereas rs174602 was inversely associated with DHA. |
| Scholtz et al (2015)29 (USA) | Pregnant women, RBC phospholipids; 8–20 wk gestation (intervention study, n = 205) | FADS1 rs174553; FADS2 rs174575 |
|
Minor allele frequency for each SNP shown in Table 6.
Abbreviations: AA, arachidonic acid; ALA, α-linolenic acid; DGLA, dihomo-γ-linolenic acid; DHA, docosahexaenoic acid; DPA, docosapentaenoic acid; EPA, eicosapentaenoic acid; FA, fatty acid; FADS, fatty acid desaturase; LA, linoleic acid; LCPUFA, long-chain polyunsaturated fatty acid; PUFA, polyunsaturated fatty acid; RBC, red blood cell; RCT, randomized controlled trial; SNP, single nucleotide polymorphism.
FADS genotype and PUFA status of breast milk
Breast milk provides the optimum nutritional composition for infants, furthermore, it is a rich source of PUFA, with the maternal diet influencing the PUFA profile of breast milk.61 The lactating mammary gland has both D5D and D6D enzymes present,33 and therefore the FADS genotype may influence PUFA concentrations of breast milk. A total of 7 studies, all of which were observational, were identified, and these studies investigated the relationship between maternal FADS genotype and breast-milk PUFA composition (Table 3). Lower breast-milk AA concentrations in carriers of the minor allele were seen in the studies identified, with the SNPs rs1535, rs174547, rs174556, rs174537, rs174570, rs2072114, rs174602, rs526126, rs174626 and rs174464, rs174546, rs174553, rs99780, and rs174583 showing such associations. Higher precursor fatty acid concentrations were also commonly found in minor allele carriers. Similar to blood PUFA status during pregnancy, the presence of the minor allele was associated with lower AA, and higher PUFA precursor, concentrations as well as lower product:substrate ratios in breast milk. The lower product:substrate ratios also suggest impaired functionality of the desaturase enzymes as a result of variation in FADS genotype. PUFA composition of breast milk appears to be influenced by FADS genotype. The evidence, however, for increased dietary intake of preformed LCPUFA for increasing breast-milk LCPUFA is not as clear. Of the 7 relevant studies included in this review, only 1 reported the influence of diet on breast-milk PUFA according to genotype, and this study investigated the effect of maternal fish and fish oil intake on DHA proportions in plasma and breast milk.32 In women who were homozygous for the minor allele, lower proportions of DHA were observed in milk and DHA concentrations were not compensated for by increased intake of fish or fish oil; however, an increase in DHA was seen in major allele homozygotes.32 Further research designed to take FADS genotype into account is needed to investigate the impact of maternal dietary PUFA on breast-milk concentrations according to FADS genotype, and to determine whether minor allele carriers would benefit from increased preformed LCPUFA intake.
Table 3.
Associations between FADS genotype and breast-milk PUFA status
| Reference (country) | Participants (study type, n) | FADS, SNP | Findings |
|---|---|---|---|
| Lattka et al (2011)39 (Germany) | Lactating mothers (observational study, n = 772) | FADS1 rs174547, rs174556; FADS2 rs174602, rs498793, rs526126; intergenic FADS2/3 rs174626; FADS3 rs1000778, rs174455 | Minor allele carriers for rs174547 and rs174556 had lower concentrations of AA and AA:DGLA ratio in breast milk than major allele carriers. |
| Moltó-Puigmartí et al (2010)32 (the Netherlands) | Lactating mothers (observational study, n = 309) | FADS1 rs174561; FADS2 rs174575; intergenic rs3834458; haplotypes for FADS1/FADS2 | Higher LA and ALA, and lower AA and DHA, proportions in breast milk in minor allele carriers than in major allele carriers. Haplotype analysis results were similar to results for single SNPs, with homozygous minor alleles having lower concentrations of product molecules such as AA compared to major allele homozygotes. |
| Mychaleckyj et al (2018)62 (Bangladesh) | Mothers (observational study, n = 1142) | FADS1 rs174556 | Major allele carriers had increased AA concentrations compared to minor allele carriers. |
| Morales et al (2011)40 (Spain) | Mother-child pairs (birth cohort study, n = 270) | FADS1 rs174537; FADS2 rs968567, rs2072114, rs526126, rs174626, rs174627; FADS3 rs174464, rs174468 | Mothers who were minor allele carriers had lower levels of AA in colostrum for SNPs rs174537, rs174570, rs2072114, rs174602, rs526126, rs174626, and rs174464. Major allele carriers for SNP rs174468 were associated with lower levels of AA. Minor allele carriers for rs174602 and rs174464 were associated with lower levels of DHA in colostrum. |
| Muc et al (2015)38 (Denmark) | Mother-child pairs (birth cohort study, n = 109) | FADS1 rs174546, rs174556 | Mothers who were minor allele carriers for rs174546 and rs174556 had lower breast-milk AA concentrations. Breast-milk EPA concentrations were also lower in minor allele carriers for rs174546. |
| Xie and Innis (2008)33 (Canada) | Lactating women (observational study, n = 54) | FADS1 intergenic rs174553; FADS2 rs99780, rs174575, rs174583; haplotypes for FADS1/FADS2 | Minor allele homozygote mothers for rs174553, rs99780, and rs174583 had lower AA, EPA, and DPA breast-milk concentrations. Minor allele homozygotes for rs174575, rs99780, and rs174583 also had lower DHA breast-milk concentrations. |
| Ding et al (2016)37 (China) | Lactating women (observational study, n = 209) | FADS1 rs174547, rs174553; FADS2 rs3834458, rs1535, rs174575, rs174602, rs498793; FADS3 rs174450, rs1000778, rs7115739; haplotypes for FADS1/FADS2 | Minor allele homozygotes for rs174547 and rs1535 had lower AA concentrations than major allele homozygotes. Rs1535 minor allele homozygotes also had lower GLA concentrations than major allele homozygotes. Higher concentrations of LA and ALA were reported in heterozygotes for rs1000778 compared to homozygous major allele carriers. Minor allele haplotypes were associated with lower concentrations of AA. |
Minor allele frequency for each SNP shown in Table 6.
Abbreviations: AA, arachidonic acid; ALA, α-linolenic acid; DGLA, dihomo-γ-linolenic acid; DHA, docosahexaenoic acid; DPA, dipicolonic acid; EPA, eicosapentaenoic acid; FADS, fatty acid desaturase; GLA, γ-linolenic acid; LA, linoleic acid; PUFA, polyunsaturated fatty acid; SNP, single nucleotide polymorphism.
FADS genotype and child PUFA status
A total of 12 studies investigating child FADS genotype and PUFA status were identified (Table 4) in subjects from birth (via cord blood) to age 13 years. At birth, PUFA status of cord blood was measured by Barman et al,41 Lattka et al,42 and Steer et al.43 It was consistently found that children with the minor allele for FADS SNPs had decreased circulating AA concentrations. Similarly, in children aged 2–13 years, children with minor allele homozygosity for FADS SNPs were observed to have lower AA concentrations.41,44–46 An association of minor allele homozygosity with DHA status – specifically, an increase in DHA status for rs1535, and a decrease for rs174448 and rs174575, when compared with major allele homozygotes – has also been reported in a Danish population.47 These associations with DHA have not been consistently shown in other studies also completed in Denmark.46,48
Table 4.
Associations between FADS genotype and child PUFA status
| Reference (country) | Participants, blood fraction for PUFA analysis (study type, n) | Maternal or child genotype; FADS, SNP | Findings |
|---|---|---|---|
| Andersen et al (2017)48 (Denmark) | Children, erythrocyte DHA; age 9 mo (prospective cohort study, n = 166) | Child; FADS2 rs1535, rs174575; intergenic rs174448 | No association was found between DHA status and FADS genotype. |
| Barman et al (2015)41 (Sweden) | Children, serum phospholipids; birth and age 13 y (birth cohort study, n = 211) | Child; intergenic rs102275, rs174448; haplotype of rs102275-rs174448 | rs102275 and rs174448 minor allele carriers had decreased AA and increased DGLA proportions at birth. The ratio of AA:DGLA was also associated with minor alleles for both SNPs. At age 13 y, lower AA proportions were reported for minor allele carriers of rs102275, and a decrease in the AA:DGLA ratio was also reported for minor allele carriers of rs102275 and rs174448. |
| Fahmida et al (2015)44 (Indonesia) | Children, plasma; age 2 y (baseline data from RCT, n = 206) | Child; FADS3 rs174468 | rs174468 heterozygous carriage was significantly associated with increased concentrations of AA, total n-6 LCPUFA, ETA, and AA:DGLA ratio, compared with major allele homozygotes. |
| Glaser et al (2011)63 (Germany) | Children, serum glycerophospholipids; age 2 and 6 y (birth cohort study, n = 331) | Child; FADS1 rs174545, rs174546, rs174556, rs174561; FADS2 rs3834458 | Higher AA and DPA (n-6) concentrations in heterozygotes and minor allele homozygotes were compared with major allele homozygotes for the SNPs analyzed. Minor allele carriers had lower concentrations of n-3 LCPUFA than those who were major allele homozygotes; however, there was no consistent trend for individual n-3 LCPUFA. |
| Harslof et al (2013)47 (Denmark) | Infants, RBC; age 9 mo, and 3 y (cross-sectional study, n = 409) | Child; FADS2 rs1535, rs174575; intergenic FADS2–FADS3 rs174448; intergenic FADS1–FADS2 rs3834458; haplotypes for rs1535, rs174575, rs174448 | At age 9 mo, minor allele homozygosity for rs1535 was associated with increased DHA status, whereas homozygosity for minor alleles of rs174448 and rs174575 was associated with decreased DHA status when compared with major allele homozygotes. For all SNPs there was increased DGLA in homozygous minor allele carriers. Minor allele carriage of rs1535 was associated with increased ALA concentrations. The AA:DGLA ratio decreased with increasing number of minor alleles for all SNPs. When children were age 3 y, there were no associations between DHA and any of the FADS SNPs genotyped. Haplotypes were not significantly associated with DHA status at 9 mo. DGLA status increased with increasing number of minor alleles. |
| Lattka et al (2013)42 (England) | Infants, plasma; cord blood collected at delivery (longitudinal study, n = 2035) | Mother and child; FADS1 rs174548, rs174556, rs174561; intergenic rs3834458, rs968567; FADS2 rs174570, rs174574, rs2727271, rs174576, rs174578, rs174579, rs174602, rs498793, rs526126; intergenic rs174448, rs174449; FADS3 rs174455; haplotypes | The minor allele was significantly associated with higher amounts of cord blood LA for all SNPs for both mothers and children, except for rs498793, where this did not reach significance, and child rs526126 was also not significant. Higher cord levels of eicosadienoic acid and DGLA, and lower amounts of AA, adrenic acid, and DPA, were also reported for maternal minor allele carriers for rs174548, rs174556, rs174561, rs3834458, rs174574, rs174576, rs174578, and rs526126. This was also found in children who were minor allele carriers for SNPs rs174548, rs174556, rs174561, rs3834458, rs174570, rs174574, rs2727271, rs174576, rs174578, rs174448, rs174449, and rs174455. Presence of the minor allele in mothers was generally associated with lower concentrations of DHA in cord blood except for rs498793, which was associated with higher levels of DHA in cord blood. Children who were carriers of the minor allele for rs498793 had lower ALA concentrations, whereas results for all other SNPs showed associations with increased ALA. In both mothers and children, minor allele carriage for all SNPs was associated with lower cord PUFA AA:DGLA and EPA:ALA ratios, except for rs498793, where the minor allele was associated with increased cord ratios. Both maternal and child FADS haplotypes influenced cord plasma LCPUFA. |
| Lupu et al (2015)64 (USA) | Toddlers, plasma; age 16 mo (observational study, n = 65) | Mother and child; FADS2 rs174575 | Maternal genotype correlated with toddler plasma ALA and AA. ALA was associated with maternal genotype in girls only, whereas AA was only associated with maternal genotype in boys. Toddler PUFA status was not correlated with toddler genotype. |
| Steer et al (2012)43 (England) | Children, plasma; cord blood at delivery; age 7 y (longitudinal study, n = 5632) | Mother and child FADS2 rs174575, rs1535 | rs174575 and rs1535 minor allele carriage in both mothers and children was associated with increased LA, and decreased AA and DHA, in cord plasma. Maternal minor allele carriers of both SNPs were associated with increased LA, and decreased AA, EPA, DPA, and DHA, in plasma from children aged 7 y. Child minor allele carriage for both SNPs was associated with increased LA, EDA, and DGLA, and decreased GLA, AA, DTA, BDPA, EPA, DPA, and DHA, in plasma of children aged 7 y. Interactions with gender were also examined. Similar fatty acid levels were found in homozygous minor allele boys and girls, but differences were seen between boys and girls for major allele carriers. Minor allele homozygote girls for rs174575 had lower DHA status than boys who were homozygous for the minor allele. |
| Rzehak et al (2010)45 (The Netherlands, Germany) | Children, plasma phospholipid, plasma glycerophospholipids (cohort study, n = 879) | Child; FADS1 rs174545, rs174546, rs174556, rs174561; FADS2 rs3834458 | All SNPs were significantly associated with all fatty acids except for ALA and EPA. |
| Wolters et al (2017)65 (Belgium, Cyprus, Estonia, Germany, Hungary, Italy, Spain, Sweden) | Children, whole blood; age 2–10 y (observational study, n = 520) | Child; FADS1 rs174546 | Higher DGLA and lower AA and EPA levels were reported for minor allele carriers of rs174546 as well as a lower D5D activity, as indicated by D5D index. |
| Lauritzen et al (2017)46 (Denmark) | Children, whole blood; age 8–11 y (cross-sectional study, n = 765) | Child; FADS2 rs1535, rs174448; FADS3 rs174468 | No significant associations were found between whole-blood DHA in schoolchildren. Minor allele carriage of rs1535 was associated with significantly lower AA concentrations compared with those who were major allele homozygous. For rs174448, minor allele carriers showed a trend towards decreased DHA status compared with major allele carriers. |
| Meldrum et al (2017)66 (Australia) | Children, plasma, erythrocyte membrane; age 6 mo (RCT, n = 259) | Child; FADS1 rs174545, rs174546, rs174548, rs174553, rs174556, rs174537; intergenic - FADS1/FADS2 rs174448, rs174449; FADS3 rs174455; haplotypes for FADS1/FADS2 | Participants in the fish oil intervention who had FADS1 minor allele haplotype were associated with higher erythrocyte DHA concentrations. Minor allele homozygotes of all SNPs of the FADS1 gene had significantly higher DHA concentrations for those in the fish oil group only. |
Minor allele frequency for each SNP shown in Table 6.
Abbreviations: AA, arachidonic acid; ALA, α-linolenic acid; BDPA, osbond acid; D5D, Δ-5 desaturase; DGLA, dihomo-γ-linolenic acid, DHA, docosahexaenoic acid; DPA, dipicolonic acid; DTA, docosapentaenoic acid; EDA, eicosadienoic; EPA, eicosapentaenoic acid; ETA, ethanolamine; FADS, fatty acid desaturase; GLA, γ-linolenic acid; LA, linoleic acid; LCPUFA, polyunsaturated fatty acid; PUFA, polyunsaturated fatty acid; RBC, red blood cell; RCT, randomized controlled trial; SNP, single nucleotide polymorphism.
During infancy, desaturase activity decreases with age, resulting in reduced synthesis of AA and DHA.67 Four studies investigated the role of child FADS genotype on PUFA product:precursor ratios. Of these, 3 studies reported that the presence of the minor allele for a range of SNPs was associated with a lower AA:DGLA ratio.42,47,65 Conversely, Fahmida et al44 found that heterozygotes for SNP 174468 had a higher AA:DGLA ratio than major allele homozygotes, indicating desaturase enzyme activity increased in the presence of genetic variation. This contrary finding may be due to the smaller sample size, and possibly also to a lower incidence of the minor allele frequency (MAF) in the Indonesian population (0.19 for rs174468) compared to the MAF of the SNPs assessed in European populations (0.14–0.81, depending on SNP).68
Three studies investigated the influence of maternal genotype on child PUFA status.42,43,64 Maternal genotype was associated with increased LA cord blood concentrations.42,43 Maternal genotype was also associated with decreased cord blood LCPUFA concentrations, specifically AA and DHA43 and lower AA:DGLA and EPA:ALA ratios.42 Interestingly, child PUFA status at age 7 years was also found to be associated with maternal genotype, with minor allele carrier mothers being associated with increased LA and decreased AA, EPA, docosapentaenoic acid (DPA), and DHA status in offspring aged 7 years.43 The associations observed between maternal genotype and child PUFA status may suggest that maternal genotype has a lasting influence on child PUFA status, or another likely explanation is that the child has inherited the maternal genotype. There may be sex differences in the interaction between FADS genotype and PUFA status. Both boys and girls were found to have similar PUFA concentrations when they were major allele homozygotes; however, in minor allele homozygotes, girls had lower DHA status than boys, and conversely minor allele homozygous boys had lower AA than girls.43 It has previously been noted that women can synthesize more DHA from ALA than men,69 thereby indicating sex and FADS genotype may interact. Future studies investigating the influence of FADS genotype on child PUFA status should consider the potential influence of sex. One randomized controlled trial investigated the influence of FADS genotype on child LCPUFA status following supplementation with either fish oil or placebo from birth to 6 months old.66 Children in the fish oil–supplemented group had significantly higher DHA status in minor allele homozygotes for all FADS1 SNPs than heterozygous and homozygous major allele carriers. This finding was not observed in children of the placebo group, suggesting that increased intake of preformed LCPUFA is beneficial for the biological status of these LCPUFAs in minor allele homozygotes. DHA and AA are both known to be essential postnatally, particularly for cognition,18 vision,17 and brain growth,5,70 and also have a role in the inflammatory response.71 DHA is known to be anti-inflammatory,71 and AA is a component of lymphocyte membrane phospholipids and CD4+ T cells, which are essential for immunoregulation.72 Given these vital roles for the immune and inflammatory response, optimal DHA and AA status in children is important, and therefore further understanding of the influence of FADS SNPs on these LCPUFA is required. Further research in the form of intervention studies is required to investigate whether increased LCPUFA intake would lead to an increase in child biological status according to genotype.
FADS genotype and child health outcomes
PUFA status is linked to several health outcomes, with increased PUFA having beneficial effects on health. Given the influence of FADS genotype on PUFA concentrations, this impact of genotype has the potential to influence health outcomes. A total of 30 studies were identified with regard to the influence of FADS genotype on child health outcomes (Table 5). These outcomes included cognition, allergies, and asthma, as well as various others that have not been as widely studied to date.
Table 5.
Associations between FADS genotype and child health outcomes
| Reference (country) | Participants (study type, n) | Maternal or child genotype; FADS, SNP | Child outcome measured | Findings |
|---|---|---|---|---|
| Caspi et al (2007)73 (New Zealand and England) | Children (birth cohort study, n = 3269) | Child; FADS2 rs174575, rs1535 | IQ | No associations between genotype and IQ. rs174575 and rs1535 interact with breastfeeding to influence IQ, with major allele carriers who were breastfed having higher IQ scores than children who were not breastfed. |
| Cheatham et al (2015)74 (USA) | Mother-child pairs; children aged 16 mo (observational, n = 71) | Maternal; FADS2 rs174575 | Declarative memory abilities | Minor allele carrier mothers for rs174575 SNP had toddlers who scored poorer in memory assessments than toddlers of homozygous major or heterozygous mothers. |
| Jensen et al (2014)75 (Denmark) | Children; age 36 mo (prospective cohort study, n = 268) | Child; FADS2 rs1535, rs174575; intergenic rs174448 | Communication, gross motor, fine motor, problem solving, and personal/social skills | There were no overall associations seen between FADS SNPs and outcomes. There were some FADS–sex interactions observed, with the minor allele of rs1535 being associated with lower communication and problem-solving scores in girls than in boys. A minor allele of rs174448 and rs174575 had the opposite effect. |
| Lauritzen et al (2017)46 (Denmark) | Children; age 8–11 y (cross-sectional study, n = 765) | Child; FADS2 rs1535, rs174448; FADS3 rs174468 | Reading test, d2 test of attention, concentration performance | rs174448 minor allele carriage was associated with lower attention and also lower reading scores in boys, but in girls the minor allele was associated with better reading scores. In girls, rs174448, the minor allele, was associated with higher reading scores, higher concentration performance, and lower attention scores compared with those who were homozygous major allele carriers. In boys, minor allele carriers for rs174448 had higher attention, lower reading scores, and lower concentration. For rs1535, minor allele–carrying girls had higher attention scores, whereas minor allele–carrying boys had lower scores. |
| Martin et al (2011)49 (Australia) | Adolescent twins; age 16 y (observational study, n = 1431) | Adolescent and maternal; FADS2 rs174575, rs174583, rs1535 | IQ | No association was found between FADS2 alleles and adolescent IQ. No significant interaction was found between FADS genotype and breastfeeding on IQ for any of the SNPs. There was no association between maternal FADS2 and child IQ at age 16 y. |
| Morales et al (2011)40 (Spain) | Mother-child pairs; children aged 14 mo and 4 y (birth cohort study, n = 740) | Maternal and child; FADS1 rs174537; FADS2 rs968567, rs2072114, rs526126, rs174626, rs174627; FADS3 rs174464, rs174468 | Cognition | Maternal minor allele carriage for SNPs rs968567, rs174627, and rs174464 was associated with higher child cognitive scores compared with major allele homozygotes. For rs174602, maternal minor allele carriers for rs174602 were associated with lower cognitive scores in children compared with children of major allele homozygous mothers. There were no significant associations between child FADS genotype and cognition. An interaction between genotype and breastfeeding was observed, with those children who were major allele homozygotes for rs174468 and had not been breastfed having lower scores for cognition. This effect was not seen in minor allele carriers for rs174468. |
| Rizzi et al (2013)76 (Belgium) | Children; twins aged 10 y (prospective study, n = 534) | Child; FADS2 rs174575, rs1535 | IQ | For rs174575 there was a trend towards higher IQ scores in breastfed children than in non-breastfed children who were major allele carriers. An interaction between genotype and breastfeeding indicated that rs1535 and breastfeeding were not significantly related to IQ; however, a larger effect was seen in minor allele carriers compared with major allele homozygotes. |
| Steer et al (2010)77 (England) | Children; age 8 y (longitudinal study, n = 5934) | Maternal and child; FADS2 rs174575, rs1535 | IQ | An interaction between breastfeeding and child genotype was associated with child IQ at age 8 y. Minor allele carriers who were formula fed scored lowest, whereas their breastfed counterparts scored similarly to breastfed children who were heterozygous or homozygous major allele carriers for both SNPs. No associations were seen between maternal genotype and child IQ, and no interaction with breastfeeding was found. |
| Steer et al (2013)78 (England) | Mother-child pairs; children aged 8 y (longitudinal study, n = 2839) | Maternal; FADS1 rs174548, rs174556, rs174561; intergenic rs3834458, rs968567; FADS2 rs174570, rs174574, rs2727271, rs174576, rs174578, rs174579, rs174602, rs498793, rs526126; intergenic rs174448 rs174449; FADS3 rs174455 | IQ | Minor allele carriage of rs3834458 was positively associated with verbal, performance and full-scale measures of IQ. There was a positive association between rs498793 and verbal measures for minor allele carriers, and also positive associations between rs968567 and performance in minor allele carriers. Negative associations were found between rs174578 and verbal measures, rs174548 and rs174455 and performance measures, and also between rs174574 and full-scale measures for those with the minor allele. The minor allele for rs968567 was associated with increased scores for performance and full-scale IQ measures. |
| Yeates et al (2015)20 (Seychelles) | Mothers and children (longitudinal study, n = 1622) | Maternal; FADS1 rs174537, rs174561; FADS1–FADS2 rs3834458; FADS2 rs174575 | Neurodevelopmental outcomes | No significant associations between maternal FADS genotype and child developmental outcomes were found. There was a trend for infants of rs3834458 minor allele–carrying mothers to score higher on the Psychomotor Developmental Index. |
| Andersen et al (2017)48 (Denmark) | Children, age 9 mo (prospective cohort study, n = 166) | Child; FADS2 rs1535, rs174575; intergenic rs174448 |
|
No significant associations were found between FADS SNPs and developmental outcomes. There was a trend towards a significant negative association between personal and social skill score and presence of the minor allele carriage for rs1535 and rs174575. For rs174575, minor allele–carrying girls had significantly higher fine motor scores than major allele homozygotes. Scores for boys did not differ between minor allele and major allele homozygous carriers. |
| Fahmida et al (2015)44 (Indonesia) | Children; age 2 y (baseline data from RCT, n = 206) | Child; FADS3 rs174468 | Cognition | No significant difference in developmental scores was found between major allele homozygotes and minor allele carriers. |
| Groen-Blokhuis et al (2013)50 (the Netherlands) | Children; twins (longitudinal study, n = 1313) | Child; FADS2 rs174575 | IQ, educational attainment, overactive behavior, attention problems | FADS gene polymorphisms were not associated with any of the cognitive outcomes, and the relationship between breastfeeding and cognitive outcomes was not moderated by FADS gene SNPs. |
| Rzehak et al (2010)45 (the Netherlands and Germany) | Children; age 2 y (cohort study, n = 879) | Child; FADS1 rs174545, rs174546, rs174556, rs174561; FADS1–FADS2 rs3834458 | Eczema | No significant associations were found between SNPs and parental reported eczema in children aged 2 y. The KOALA study found no significant associations between genotype and eczema. In the LISA study, all FADS1 FADS2 SNPs were significantly associated with eczema reported by parents in their children up to age 2 y. |
| Standl et al (2011)51 (Germany) | Children; age 10 y (prospective cohort study, n = 2000) | Child; FADS1 rs174545, rs174546, rs174556, rs174561; FADS1–FADS2 rs3834458; FADS2 rs174575 | Atopic diseases | No significant association was found between FADS SNPs and prevalence of atopic diseases or allergic sensitization. There was an association between n-6:n-3 ratio and increased risk of hayfever in major allele homozygotes for the 6 SNPs included in this study. Asthma was significantly positively associated with daily margarine intake in major allele homozygotes for each SNP. |
| Barman et al (2015)41 (Sweden) | Children; at birth and age 13 y (birth cohort study, n = 211) | Child; intergenic rs102275, rs174448 | Allergies | Minor allele carriers of rs102275 and rs174448 had reduced risk of developing atopic eczema at age 13 y compared with major allele homozygotes. No associations were found between FADS SNPs and respiratory allergy. |
| Standl et al (2012)52 (Germany) | Children; age 10 y (prospective cohort study, n = 2245) | Child; FADS1 rs174545, rs174546, rs174556, rs174561; FADS1–FADS2 rs3834458; FADS2 rs174575 | Asthma | There was a lower prevalence of asthma in minor allele carriers than in major allele homozygotes; however, this effect was not significant. For all SNPs, children with the minor allele who were exclusively breastfed for a minimum of 3 mo had a reduced prevalence of asthma; however, there was no effect in major allele homozygotes. For SNPs rs174545, rs174546, rs174556, rs174561, and rs3834458, reduction in asthma risk was greatest in those who were heterozygous or homozygous for the minor allele and who were exclusively breastfed for 3 or 4 mo after birth. The asthma risk was further reduced with longer exclusive breastfeeding for minor allele carriers of rs174575 only. |
| Standl et al (2012)53 (Germany) | Children; age 10 y (prospective birth cohort study, n = 2006) | Child; FADS1 rs174545, rs174546, rs174556, rs174561; FADS1–FADS2 rs3834458; FADS2 rs174575 | Serum lipids and lipoproteins | Minor allele homozygotes had lower total cholesterol and LDL than major allele homozygotes. Heterozygotes had lower HDL levels and higher triglyceride concentrations than major allele homozygotes. |
| Moltó-Puigmartí et al (2013)54 (the Netherlands) | Children; age 2 y (prospective birth cohort study, n = 521) | Child; FADS1 rs174545, rs174546, rs174556, rs174561; FADS1–FADS2 rs3834458 | Plasma cholesterol concentrations | For all FADS SNPs, minor allele carriers had lower total cholesterol than major allele homozygotes. Minor allele homozygotes for rs174546 and rs3834458 also had significantly lower HDLc concentrations than those who were homozygous for the major allele. |
| Costea et al (2014)79 (Canada) | Children; age <19 y (case control, n = 432) | Child; FADS2 rs11230815, rs17831757, rs968567, rs174627 | Crohn’s disease | Children with higher dietary intake of n-6:n-3 ratio who were carriers of minor allele for FADS2 SNPs were more susceptible for Crohn’s disease. |
| Hovsepian et al (2018)80 (Iran) | Children (case control, n = 528) | Child; FADS1 rs174547 | Metabolic Syndrome | In children with metabolic syndrome, there was a significantly higher prevalence of the minor allele for rs174547 than in those children without metabolic syndrome. |
| Sun et al (2018)81(China) | Children (case control, n = 486) | Child; FADS2 rs526126; haplotype for rs526126 | Autism spectrum disorder (ASD) | The minor allele for FADS2 rs526126 was associated with decreased risk of ASD. No differences in frequencies of haplotypes were found between ASD patients and controls. |
| Wolters et al (2017)65 (Belgium, Cyprus, Estonia, Germany, Hungary, Italy, Spain, Sweden) | Children; age 2–10 y (observational study, n = 520) | Child; FADS1 rs174546 | Blood pressure | The presence of the minor allele had a lowering effect on blood pressure via an interaction with AA and BMI. There was no direct effect of the minor allele on blood pressure. |
| Brookes et al (2006)82 (England) | Children and adolescents (case control, n = 360) | Child; FADS1 rs174545, rs174548; FADS2 rs498793; haplotypes | ADHD | FADS2 rs498793 was associated with ADHD. No evidence of association was found between ADHD and haplotype. |
| Golding et al (2012)83 (England) | Children; age 8 y (case control, n = 7831) | Child; FADS1 rs174548, rs174556, rs174561; intergenic FADS1–FADS2 rs3834458, rs968567; FADS2 rs174570, rs1535, rs174574, rs174575, rs2727271, rs174576, rs174578, rs174579, rs174602, rs498793, rs526126; intergenic FADS2–FADS2 rs174448, rs174449; FADS3 rs174455 | Growing pains | No associations were found between FADS genotype and incidence of reported growing pains. |
| Muc et al (2015)38 (Denmark) | Mother-child pairs (birth cohort study, n = 109) | Maternal and child; FADS1 rs174546, rs174556 | Inflammatory markers | Maternal FADS SNP was negatively correlated with all cytokines analyzed in infants, with correlations being significant for IL-17 for rs174546. There were no significant correlations between any of the cytokines and child FADS genotype. |
| Moltó-Puigmartí et al (2014)84 (the Netherlands) | Mother-child pairs (prospective birth cohort study, n = 2669) | Maternal and child; FADS1 rs174556 | Pregnancy duration and birthweight | Women who were minor allele homozygotes had significantly shorter pregnancies. There was no association between pregnancy duration and fetal genotype. Minor allele homozygous mothers had lighter children than major allele homozygote mothers. There was no significant association between fetal genotype and birthweight. |
| Bernard et al (2018)85 (Singapore) | Mothers and their offspring (birth cohort study, n = 898 mothers, n = 1103 children) | Maternal and child; FADS1 rs174546, rs174547, rs174548, rs174550; FADS2 rs174570, rs1535, rs2727270, rs174576, rs174577, rs174583, rs2851682, rs174593, rs498793, rs174611, rs174618, rs17156506; FADS3 rs174450, rs1000778, rs174455 | Gestation duration and birth size | Both offspring and maternal FADS3 SNPs were associated with gestation duration. The minor allele for rs174450 in both mothers and offspring was associated with shorter gestation. Minor allele homozygote mothers for rs174450 also had quicker delivery after spontaneous labor compared with major allele homozygotes. |
| Liu et al (2012)86 (USA) | Mothers (observational study, n = 1110) | Maternal; FADS1 rs2072114, rs7119667 | Preterm delivery | Minor allele carriers of rs2072114 and rs7119667 were more likely to have a preterm delivery. |
| Gonzalez-Casanova et al (2016)28 (Mexico) | Women and their offspring (RCT, n = 654) | Maternal; FADS1 rs174556; FADS2 rs174602, rs498793; FADS3 rs174455 | Birth weight | Children whose mothers were minor allele carriers for rs174602 and were in the DHA-supplemented group were heavier than children whose mothers were in the placebo group. There were no associations between birthweight and any of the other SNPs. |
Minor allele frequency for each SNP shown in Table 6.
Abbreviations: AA, arachidonic acid; ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder; BMI, body mass index; DHA, docosahexaenoic acid; FADS, fatty acid desaturase; HDL, high-density lipoprotein; HDLc, high-density lipoprotein cholesterol; IL-17, interleukin-17; IQ, intelligence quotient; LDL, low-density lipoprotein; RCT, randomized controlled trial; SNP, single nucleotide polymorphism.
One of the most commonly investigated outcomes in relation to child FADS genotype is child cognition, with the current review identifying 13 relevant studies. All the included studies relating to cognition are observational in study design. Of these studies, some have reported no association between child FADS genotype and cognitive outcomes.40,44,48–50 Nevertheless, when breastfeeding was also taken into consideration, being breastfed was shown to modify the effect of genotype on child cognition. Children who were major allele carriers and breastfed had higher intelligence quotient scores than major allele children who were not breastfed.73,76 In addition to interactions with child PUFA status, sex of the child has also been found to be involved in the interaction between child genotype and child cognition. Carriage of the minor allele for rs174448 was associated with lower reading scores in boys, but improved reading scores in girls.46 Furthermore, in girls, minor allele carriage of rs174575 was associated with increased fine motor skills scores compared with major allele carriers, whilst in boys no difference in scores was observed between major and minor allele carriers.
Whilst some studies have examined the influence of maternal FADS genotype on child outcomes, the research to date has not shown a conclusive association between maternal FADS genotype and child cognition. Maternal minor allele carriage for a range of FADS SNPs has been found to be associated with improved cognitive scores, with both positive40,78 and negative associations being reported,40,74,78 depending on the FADS SNP being investigated. In contrast to this observation, other studies have found no associations between maternal FADS and child cognition after controlling for other confounders.20,50,77
The association between child FADS genotype and allergy development in children has also been investigated. Observational evidence to date has had varied outcomes. Minor alleles of FADS SNPs were associated with an increase in eczema in children aged 2 years in the LISA study45 but a decreased risk of eczema in a study conducted in 13-year-olds in Sweden.41 When the children of the LISA birth cohort study were followed up again at age 10 years, an association between FADS minor alleles and eczema was no longer reported.51 Some studies have found no overall association between FADS genotype and allergy development; however, when diet-gene interactions are taken into consideration, an association between genotype and allergies has been shown. In major allele homozygotes, an increased n-6:n-3 dietary intake has been associated with increased risk of hay fever, and higher margarine consumption with increased risk of asthma.51 Furthermore, children with the minor allele for FADS SNPs who were exclusively breastfed had a reduced prevalence of asthma, whereas no association of breastfeeding was observed in major allele homozygotes.52 This observation highlights that dietary intake may also play an important role in the association between FADS genotype and allergies. Further studies are required to investigate this association, perhaps in populations with a high PUFA status.
Maternal FADS genotype has been linked to birthweight and preterm delivery, with observational studies indicating minor allele carriers had lighter babies and shorter pregnancies. To determine conclusively the influence of FADS genotype (both maternal and child) on child outcomes, further observational and intervention studies specifically designed to assess this in different populations are required. Such future studies should also take into consideration maternal PUFA status and FADS, as well as child PUFA status, child FADS, and sex of the child, as these too may influence outcomes.
SNPs that have been commonly assessed in multiple studies for child health outcomes are shown in Table S4 in the Supporting Information online. It is evident that the results for a SNP are not necessarily consistent across studies. The association between SNP rs1535 and cognition, for example, demonstrates such mixed results. As indicated in Table S4 in the Supporting Information online, Andersen et al48 and Martin et al49 report no association between rs1535 and child neurodevelopmental outcomes; however, an interaction between rs1535 and breastfeeding,73,76,77 or rs1535 and sex of the child,46,75 has been observed. These inconsistencies have also been reported in a range of other SNPS (see Table S4 in the Supporting Information online). It is also possible that these inconsistencies may be due to differing MAFs according to ethnicity. For example, the prevalence of SNP rs3834458 varies across different population groups, with an African population having a MAF of 0.021 but a European population having a MAF of 0.346 (Table 6). The mixed results for associations between FADS genotype and PUFA status or child outcomes may also be related to the distinct cultural differences in prevalence.
Table 6.
Minor allele frequency for SNPs included in current review
| SNP | Global MAF | African | East Asian | European | South Asian | American |
|---|---|---|---|---|---|---|
| rs1000778 | 0.523 | 0.221 | 0.686 | 0.729 | 0.610 | 0.440 |
| rs102275 | 0.493 | 0.697 | 0.567 | 0.363 | 0.160 | 0.650 |
| rs11230815 | 0.932 | 0.918 | 0.987 | 0.880 | 0.970 | 0.900 |
| rs1474553 | 0.455 | 0.053 | 0.693 | 0.607 | 0.570 | 0.500 |
| rs1535 | 0.322 | 0.109 | 0.566 | 0.350 | 0.140 | 0.590 |
| rs17156506 | 0.057 | 0.141 | 0.062 | 0.001 | 0.030 | 0.010 |
| rs174448 | 0.596 | 0.576 | 0.697 | 0.616 | 0.650 | 0.380 |
| rs174449 | 0.515 | 0.343 | 0.681 | 0.619 | 0.580 | 0.370 |
| rs174450 | 0.435 | 0.207 | 0.607 | 0.528 | 0.560 | 0.310 |
| rs174455 | 0.407 | 0.033 | 0.587 | 0.614 | 0.570 | 0.330 |
| rs174464 | 0.519 | 0.211 | 0.680 | 0.730 | 0.610 | 0.440 |
| rs174468 | 0.163 | 0.024 | 0.030 | 0.409 | 0.190 | 0.220 |
| rs174537 | 0.303 | 0.025 | 0.567 | 0.349 | 0.160 | 0.590 |
| rs174545 | 0.298 | 0.022 | 0.566 | 0.347 | 0.140 | 0.590 |
| rs174546 | 0.298 | 0.022 | 0.566 | 0.347 | 0.140 | 0.590 |
| rs174547 | 0.298 | 0.023 | 0.566 | 0.347 | 0.140 | 0.590 |
| rs174548 | 0.327 | 0.179 | 0.548 | 0.314 | 0.130 | 0.590 |
| rs174550 | 0.298 | 0.022 | 0.566 | 0.347 | 0.140 | 0.590 |
| rs174553 | 0.297 | 0.021 | 0.565 | 0.347 | 0.140 | 0.590 |
| rs174556 | 0.720 | 0.020 | 0.547 | 0.303 | 0.130 | 0.570 |
| rs174561 | 0.280 | 0.020 | 0.547 | 0.303 | 0.130 | 0.570 |
| rs174570 | 0.228 | 0.009 | 0.556 | 0.160 | 0.070 | 0.490 |
| rs174574 | 0.530 | 0.367 | 0.433 | 0.638 | 0.860 | 0.360 |
| rs174575 | 0.209 | 0.210 | 0.166 | 0.255 | 0.100 | 0.360 |
| rs174576 | 0.363 | 0.246 | 0.568 | 0.358 | 0.140 | 0.610 |
| rs174577 | 0.392 | 0.346 | 0.568 | 0.361 | 0.140 | 0.620 |
| rs174578 | 0.393 | 0.349 | 0.568 | 0.363 | 0.140 | 0.620 |
| rs174579 | 0.135 | 0.016 | 0.152 | 0.203 | 0.080 | 0.320 |
| rs174583 | 0.369 | 0.242 | 0.580 | 0.363 | 0.150 | 0.610 |
| rs174593 | 0.207 | 0.184 | 0.156 | 0.264 | 0.130 | 0.360 |
| rs174602 | 0.423 | 0.741 | 0.365 | 0.213 | 0.200 | 0.520 |
| rs174611 | 0.116 | 0.045 | 0.015 | 0.286 | 0.080 | 0.200 |
| rs174618 | 0.349 | 0.597 | 0.028 | 0.432 | 0.290 | 0.300 |
| rs174626 | 0.481 | 0.350 | 0.667 | 0.536 | 0.520 | 0.320 |
| rs174627 | 0.054 | 0.021 | 0.000 | 0.145 | 0.040 | 0.090 |
| rs17831757 | 0.057 | 0.040 | 0.014 | 0.119 | 0.030 | 0.100 |
| rs2072114 | 0.195 | 0.114 | 0.417 | 0.150 | 0.060 | 0.280 |
| rs2727270 | 0.163 | 0.006 | 0.417 | 0.144 | 0.060 | 0.260 |
| rs2727271 | 0.163 | 0.006 | 0.417 | 0.144 | 0.060 | 0.270 |
| rs2851682 | 0.149 | 0.003 | 0.412 | 0.106 | 0.050 | 0.250 |
| rs3834458 | 0.296 | 0.021 | 0.566 | 0.346 | 0.130 | 0.580 |
| rs498793 | 0.687 | 0.637 | 0.918 | 0.569 | 0.690 | 0.620 |
| rs526126 | 0.680 | 0.238 | 0.824 | 0.811 | 0.880 | 0.840 |
| rs7115739 | 0.759 | 0.693 | 0.717 | 0.962 | 0.780 | 0.620 |
| rs7119667 | 0.024 | 0.086 | 0.000 | 0.000 | 0.000 | 0.010 |
| rs968567 | 0.053 | 0.011 | 0.000 | 0.150 | 0.040 | 0.080 |
| rs99780 | 0.397 | 0.374 | 0.566 | 0.358 | 0.140 | 0.620 |
Global MAF according to “1000Genomes” data available from dbSNP at www.ncbi.nlm.nih.gov.68
Abbreviations: MAF, minor allele frequency; SNP, single nucleotide polymorphism.
CONCLUSION
In conclusion, FADS genotype has an influence on the PUFA status of pregnant women, breast milk, and children, as well as being linked to some child development and health outcomes. Minor allele carriers have consistently been shown to have lower PUFA status than major allele homozygotes, likely owing to the impaired efficiency of the elongation pathway observed with FADS genotype variation. There has been a noted imbalance in dietary intake of n-6 and n-3 PUFA.87 This may also contribute to the issues in PUFA metabolism in those with the minor alleles given that both the n-6 and n-3 precursors compete for the same desaturase enzymes. It is not clear whether increased dietary intake of preformed LCPUFA will increase biological status, so further studies are needed to investigate the diet-gene interaction. Future studies should also focus on the influence of FADS on child health outcomes to explore the role of genotype on this, taking into consideration all factors, including PUFA status and FADS genotype of both the mother and child, as well as sex of the child, which may impact outcomes.
Supplementary Material
Acknowledgments
Author contributions. M.C.C. was responsible for developing, and subsequently conducting, the search strategies. Titles and abstracts were screened and evaluated by M.C.C. M.C.C also prepared the manuscript. M.C.C., E.M.M., M.S.M., J.J.S., E.vW., and A.J.Y. reviewed the manuscript.
Funding. M.C.C is a recipient of Department of Employment and Learning (DEL) postgraduate studentship. DEL had no role in the preparation of this manuscript. This study was supported by the US National Institutes of Health (grant R01-ES010219).
Declaration of interest. The authors have no relevant interests to declare.
Supporting Information
The following Supporting Information is available through the online version of this article at the publisher’s website.
Appendix S1 PRISMA checklist and search strategy for online databases
Table S1 Quality assessment of case-control studies
Table S2 Quality assessment of cohort studies
Table S3 Assessment of risk of bias for intervention studies using the Cochrane Collaboration tool for assessing risk of bias
Table S4 Summary table of associations between FADS SNPs assessed in multiple studies and child outcomes
References
- 1. Calder PC. Functional roles of fatty acids and their effects on human health. JPEN J Parenter Enteral Nutr. 2015;39(1 suppl):8S–32S. [DOI] [PubMed] [Google Scholar]
- 2. Lattka E, Illig T, Koletzko B, et al. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol. 2010;21:64–69. [DOI] [PubMed] [Google Scholar]
- 3. Uauy RD, Birch DG, Birch EE, et al. Effect of dietary omega-3 fatty acids on retinal function of very-low-birth-weight neonates. Pediatr Res. 1990;28:485–492. [DOI] [PubMed] [Google Scholar]
- 4. Xiang M, Alfven G, Blennow M, et al. Long-chain polyunsaturated fatty acids in human milk and brain growth during early infancy. Acta Paediatr. 2007;89:142–147. [DOI] [PubMed] [Google Scholar]
- 5. Tallima H, El Ridi R.. Arachidonic acid: physiological roles and potential health benefits—a review. J Adv Res. 2018;11:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ryan AS, Astwood JD, Gautier S, et al. Effects of long-chain polyunsaturated fatty acid supplementation on neurodevelopment in childhood: a review of human studies. Prostaglandins Leukot Essent Fatty Acids. 2010;82:305–314. [DOI] [PubMed] [Google Scholar]
- 7. Katsuki H, Okuda S.. Arachidonic acid as a neurotoxic and neurotrophic substance. Prog Neurobiol. 1995;46:607–636. [DOI] [PubMed] [Google Scholar]
- 8. Dobbing J, Sands J.. Comparative aspects of the brain growth spurt. Early Hum Dev. 1979;3:79–83. [DOI] [PubMed] [Google Scholar]
- 9. Fukaya T, Gondaira T, Kashiyae Y, et al. Arachidonic acid preserves hippocampal neuron membrane fluidity in senescent rats. Neurobiol Aging. 2007;28:1179–1186. [DOI] [PubMed] [Google Scholar]
- 10. Wang ZJ, Liang CL, Li GM, et al. Neuroprotective effects of arachidonic acid against oxidative stress on rat hippocampal slices. Chem Biol Interact. 2006;163:207–217. [DOI] [PubMed] [Google Scholar]
- 11. Middleton P, Gomersall JC, Gould JF, et al. Omega‐3 fatty acid addition during pregnancy. Cochrane Database Syst Rev. 2018;11:CD003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Best KP, Gold M, Kennedy D, et al. Omega-3 long-chain PUFA intake during pregnancy and allergic disease outcomes in the offspring: a systematic review and meta-analysis of observational studies and randomized controlled trials. Am J Clin Nutr. 2016;103:128–143. [DOI] [PubMed] [Google Scholar]
- 13. Bisgaard H, Stokholm J, Chawes BL, et al. Fish oil-derived fatty acids in pregnancy and wheeze and asthma in offspring. N Engl J Med. 2016;375:2530–2539. [DOI] [PubMed] [Google Scholar]
- 14. Hidaka BH, Thodosoff JM, Kerling EH, et al. Intrauterine DHA exposure and child body composition at 5 y: exploratory analysis of a randomized controlled trial of prenatal DHA supplementation. Am J Clin Nutr. 2018;107:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kerling EH, Hilton JM, Thodosoff JM, et al. Effect of prenatal docosahexaenoic acid supplementation on blood pressure in children with overweight condition or obesity: a secondary analysis of a randomized clinical trial. JAMA Netw Open. 2019;2:e190088.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gould JF, Smithers LG, Makrides M.. The effect of maternal omega-3 (n-3) LCPUFA supplementation during pregnancy on early childhood cognitive and visual development: a systematic review and meta-analysis of randomized controlled trials. Am J Clin Nutr. 2013;97:531–544. [DOI] [PubMed] [Google Scholar]
- 17. Hoffman DR, Boettcher JA, Diersen-Schade DA.. Toward optimizing vision and cognition in term infants by dietary docosahexaenoic and arachidonic acid supplementation: a review of randomized controlled trials. Prostaglandins Leukot Essent Fatty Acids. 2009;81:151–158. [DOI] [PubMed] [Google Scholar]
- 18. Birch EE, Garfield S, Hoffman DR, et al. A randomized controlled trial of early dietary supply of long-chain polyunsaturated fatty acids and mental development in term infants. Dev Med Child Neurol. 2000;42:174–181. [DOI] [PubMed] [Google Scholar]
- 19. Lattka E, Illig T, Heinrich J, et al. FADS gene cluster polymorphisms: important modulators of fatty acid levels and their impact on atopic diseases. J Nutrigenet Nutrigenomics. 2009;2:119–128. [DOI] [PubMed] [Google Scholar]
- 20. Yeates AJ, Love TM, Engstrom K, et al. Genetic variation in FADS genes is associated with maternal long-chain PUFA status but not with cognitive development of infants in a high fish-eating observational study. Prostaglandins Leukot Essent Fatty Acids. 2015;102–103:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ameur A, Enroth S, Johansson A, et al. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet. 2012;90:809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vessby B, Gustafsson IB, Tengblad S, et al. Indices of fatty acid desaturase activity in healthy human subjects: effects of different types of dietary fat. Br J Nutr. 2013;110:871–879. [DOI] [PubMed] [Google Scholar]
- 23. Calder PC. Docosahexaenoic acid. Ann Nutr Metab. 2016;69(suppl 1):8–21. [DOI] [PubMed] [Google Scholar]
- 24. Simopoulos AP. The omega-6/omega-3 fatty acid ratio: health implications. OCL. 2010;17:267–275. [Google Scholar]
- 25. Gregory MK, Gibson RA, Cook-Johnson RJ, et al. Elongase reactions as control points in long-chain polyunsaturated fatty acid synthesis. PLoS One. 2011;6:e29662.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carlier H, Bernard A, Caselli C.. Digestion and absorption of polyunsaturated fatty acids. Reprod Nutr Dev. 1991;31:475–500. [DOI] [PubMed] [Google Scholar]
- 27. de la Garza Puentes A, Montes Goyanes R, Chisaguano Tonato AM, et al. Association of maternal weight with FADS and ELOVL genetic variants and fatty acid levels – the PREOBE follow-up. PLoS One. 2017;12:e0179135.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gonzalez-Casanova I, Rzehak P, Stein AD, et al. Maternal single nucleotide polymorphisms in the fatty acid desaturase 1 and 2 coding regions modify the impact of prenatal supplementation with DHA on birth weight. Am J Clin Nutr. 2016;103:1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scholtz SA, Kerling EH, Shaddy DJ, et al. Docosahexaenoic acid (DHA) supplementation in pregnancy differentially modulates arachidonic acid and DHA status across FADS genotypes in pregnancy. Prostaglandins Leukot Essent Fatty Acids. 2015;94:29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koletzko B, Lattka E, Zeilinger S, et al. Genetic variants of the fatty acid desaturase gene cluster predict amounts of red blood cell docosahexaenoic and other polyunsaturated fatty acids in pregnant women: findings from the Avon Longitudinal Study of Parents and Children. Am J Clin Nutr. 2011;93:211–219. [DOI] [PubMed] [Google Scholar]
- 31. Xie L, Innis SM.. Association of fatty acid desaturase gene polymorphisms with blood lipid essential fatty acids and perinatal depression among Canadian women: a pilot study. J Nutrigenet Nutrigenomics. 2009;2:243–250. [DOI] [PubMed] [Google Scholar]
- 32. Moltó-Puigmartí C, Plat J, Mensink RP, et al. FADS1 FADS2 gene variants modify the association between fish intake and the docosahexaenoic acid proportions in human milk. Am J Clin Nutr. 2010;91:1368–1376. [DOI] [PubMed] [Google Scholar]
- 33. Xie L, Innis SM.. Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n-6) and (n-3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. J Nutr. 2008;138:2222–2228. [DOI] [PubMed] [Google Scholar]
- 34. Brenna JT, Kothapalli KS, Park WJ.. Alternative transcripts of fatty acid desaturase (FADS) genes. Prostaglandins Leukot Essent Fatty Acids. 2010;82:281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rahbar E, Ainsworth HC, Howard TD, et al. Uncovering the DNA methylation landscape in key regulatory regions within the FADS cluster. PloS One. 2017;12:e0180903.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ralston JC, Matravadia S, Gaudio N, et al. Polyunsaturated fatty acid regulation of adipocyte FADS1 and FADS2 expression and function. Obesity. 2015;23:725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ding Z, Liu G, Li X, et al. Association of polyunsaturated fatty acids in breast milk with fatty acid desaturase gene polymorphisms among Chinese lactating mothers. Prostaglandins Leukot Essent Fatty Acids. 2016;109:66–71. [DOI] [PubMed] [Google Scholar]
- 38. Muc M, Kreiner-Moller E, Larsen JM, et al. Maternal fatty acid desaturase genotype correlates with infant immune responses at 6 months. Br J Nutr. 2015;114:891–898. [DOI] [PubMed] [Google Scholar]
- 39. Lattka E, Rzehak P, Szabo E, et al. Genetic variants in the FADS gene cluster are associated with arachidonic acid concentrations of human breast milk at 1.5 and 6 mo postpartum and influence the course of milk dodecanoic, tetracosenoic, and trans-9-octadecenoic acid concentrations over the duration of lactation. Am J Clin Nutr. 2011;93:382–391. [DOI] [PubMed] [Google Scholar]
- 40. Morales E, Bustamante M, Gonzalez JR, et al. Genetic variants of the FADS gene cluster and ELOVL gene family, colostrums LC-PUFA levels, breastfeeding, and child cognition. PLoS One. 2011;6:e17181.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barman M, Nilsson S, Torinsson Naluai A, et al. Single nucleotide polymorphisms in the FADS gene cluster but not the ELOVL2 gene are associated with serum polyunsaturated fatty acid composition and development of allergy (in a Swedish birth cohort). Nutrients. 2015;7:10100–10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lattka E, Koletzko B, Zeilinger S, et al. Umbilical cord PUFA are determined by maternal and child fatty acid desaturase (FADS) genetic variants in the Avon Longitudinal Study of Parents and Children (ALSPAC). Br J Nutr. 2013;109:1196–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Steer CD, Hibbeln JR, Golding J, et al. Polyunsaturated fatty acid levels in blood during pregnancy, at birth and at 7 years: their associations with two common FADS2 polymorphisms. Hum Mol Genet. 2012;21:1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fahmida U, Htet MK, Adhiyanto C, et al. Genetic variants of FADS gene cluster, plasma LC-PUFA levels and the association with cognitive function of under-two-year-old Sasaknese Indonesian children. Asia Pac J Clin Nutr. 2015;24:323–328. [DOI] [PubMed] [Google Scholar]
- 45. Rzehak P, Thijs C, Standl M, et al. Variants of the FADS1 FADS2 gene cluster, blood levels of polyunsaturated fatty acids and eczema in children within the first 2 years of life. PLoS One. 2010;5:e13261.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lauritzen L, Sorensen LB, Harslof LB, et al. Mendelian randomization shows sex-specific associations between long-chain PUFA-related genotypes and cognitive performance in Danish schoolchildren. Am J Clin Nutr. 2017;106:88–95. [DOI] [PubMed] [Google Scholar]
- 47. Harslof LBS, Larsen LH, Ritz C, et al. FADS genotype and diet are important determinants of DHA status: a cross-sectional study in Danish infants. Am J Clin Nutr. 2013;97:1403–1410. [DOI] [PubMed] [Google Scholar]
- 48. Andersen KR, Harslof LBS, Schnurr TM, et al. A study of associations between early DHA status and fatty acid desaturase (FADS) SNP and developmental outcomes in children of obese mothers. Br J Nutr. 2017;117:278–286. [DOI] [PubMed] [Google Scholar]
- 49. Martin NW, Benyamin B, Hansell NK, et al. Cognitive function in adolescence: testing for interactions between breast-feeding and FADS2 polymorphisms. J Am Acad Child Adolesc Psychiatry. 2011;50:55–62.e4. [DOI] [PubMed] [Google Scholar]
- 50. Groen-Blokhuis MM, Franic S, van Beijsterveldt CEM, et al. A prospective study of the effects of breastfeeding and FADS2 polymorphisms on cognition and hyperactivity/attention problems. Am J Med Genet. 2013;162:457–465. [DOI] [PubMed] [Google Scholar]
- 51. Standl M, Sausenthaler S, Lattka E, et al. FADS gene variants modulate the effect of dietary fatty acid intake on allergic diseases in children. Clin Exp Allergy. 2011;41:1757–1766. [DOI] [PubMed] [Google Scholar]
- 52. Standl M, Sausenthaler S, Lattka E, et al. FADS gene cluster modulates the effect of breastfeeding on asthma. Results from the GINIplus and LISAplus studies. Allergy. 2012;67:83–90. [DOI] [PubMed] [Google Scholar]
- 53. Standl M, Lattka E, Stach B, et al. FADS1 FADS2 gene cluster, PUFA intake and blood lipids in children: results from the GINIplus and LISAplus studies. PLoS One. 2012;7:e37780.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moltó-Puigmartí C, Jansen E, Heinrich J, et al. Genetic variation in FADS genes and plasma cholesterol levels in 2-year-old infants: KOALA birth cohort study. PLoS One. 2013;8:e61671.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Non-Randomised Studies in Meta-Analyses. The Ottawa Hospital; 2011. Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed December 2018. [Google Scholar]
- 56. Higgins JPT, Green S, eds. Cochrane Handbook for Systematic Reviews of Interventions. Version 5.1. London, UK: The Cochrane Collaboration; 2011. Available at: http://handbook.cochrane.org/. Accessed December 2018. [Google Scholar]
- 57. Scholtz SA, Colombo J, Carlson SE.. Clinical overview of effects of dietary long-chain polyunsaturated fatty acids during the perinatal period. Nestle Nutr Inst Workshop Ser. 2013;77:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dutta-Roy AK. Cellular uptake of long-chain fatty acids: role of membrane-associated fatty-acid-binding/transport proteins. Cell Mol Life Sci. 2000;57:1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Herrera E. Implications of dietary fatty acids during pregnancy on placental, fetal and postnatal development – a review. Placenta. 2002;23(suppl A):S9–S19. [DOI] [PubMed] [Google Scholar]
- 60. Bonham MP, Duffy EM, Wallace JMW, et al. Habitual fish consumption does not prevent a decrease in LCPUFA status in pregnant women (the Seychelles Child Development Nutrition Study). Prostaglandins Leukot Essent Fatty Acids. 2008;78:343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ballard O, Morrow AL.. Human milk composition: nutrients and bioactive factors. Pediatr Clin North Am. 2013;60:49–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mychaleckyj JC, Nayak U, Colgate ER, et al. Multiplex genomewide association analysis of breast milk fatty acid composition extends the phenotypic association and potential selection of FADS1 variants to arachidonic acid, a critical infant micronutrient. J Med Genet. 2018;55:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Glaser C, Rzehak P, Demmelmair H, et al. , and the LISA Study Group. Influence of FADS polymorphisms on tracking of serum glycerophospholipid fatty acid concentrations and percentage composition in children. PLoS One. 2011;6:e21933.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lupu DS, Cheatham CL, Corbin KD, et al. Genetic and epigenetic transgenerational implications related to omega-3 fatty acids. Part I: maternal FADS2 genotype and DNA methylation correlate with polyunsaturated fatty acid status in toddlers: an exploratory analysis. Nutr Res. 2015;35:939–947. [DOI] [PubMed] [Google Scholar]
- 65. Wolters M, Dering C, Siani A, et al. The role of a FADS1 polymorphism in the association of fatty acid blood levels, BMI and blood pressure in young children-Analyses based on path models. PLoS One. 2017;12:e0181485.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Meldrum SJ, Li Y, Zhang G, et al. Can polymorphisms in the fatty acid desaturase (FADS) gene cluster alter the effects of fish oil supplementation on plasma and erythrocyte fatty acid profiles? An exploratory study. Eur J Nutr. 2017;58:2583–2594. [DOI] [PubMed] [Google Scholar]
- 67. Carnielli VP, Simonato M, Verlato G, et al. Synthesis of long-chain polyunsaturated fatty acids in preterm newborns fed formula with long-chain polyunsaturated fatty acids. Am J Clin Nutr. 2007;86:1323–1330. [DOI] [PubMed] [Google Scholar]
- 68.dbSNP. dbSNP short genetic variations. 2018. Available at: https://www.ncbi.nlm.nih.gov/snp/. Accessed November 2018.
- 69. Lohner S, Fekete K, Marosvolgyi T, et al. Gender differences in the long-chain polyunsaturated fatty acid status: systematic review of 51 publications. Ann Nutr Metab. 2013;62:98–112. [DOI] [PubMed] [Google Scholar]
- 70. Hadley KB, Ryan AS, Forsyth S, et al. The essentiality of arachidonic acid in infant development. Nutrients. 2016;8:216.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Calder PC. Omega-3 fatty acids and inflammatory processes. Nutrients. 2010;2:355–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Harbige LS. Fatty acids, the immune response, and autoimmunity: a question of n-6 essentiality and the balance between n-6 and n-3. Lipids. 2003;38:323–341. [DOI] [PubMed] [Google Scholar]
- 73. Caspi A, Williams B, Kim-Cohen J, et al. Moderation of breastfeeding effects on the IQ by genetic variation in fatty acid metabolism. Proc Natl Acad Sci USA. 2007;104:18860–18865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cheatham CL, Lupu DS, Niculescu MD.. Genetic and epigenetic transgenerational implications related to omega-3 fatty acids. Part II: maternal FADS2 rs174575 genotype and DNA methylation predict toddler cognitive performance. Nutr Res. 2015;35:948–955. [DOI] [PubMed] [Google Scholar]
- 75. Jensen HAR, Harslof LBS, Nielsen MS, et al. FADS single-nucleotide polymorphisms are associated with behavioral outcomes in children, and the effect varies between sexes and is dependent on PPAR genotype. Am J Clin Nutr. 2014;100:826–832. [DOI] [PubMed] [Google Scholar]
- 76. Rizzi TS, van der Sluis S, Derom C, et al. FADS2 genetic variance in combination with fatty acid intake might alter composition of the fatty acids in brain. PLoS One. 2013;8:e68000.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Steer CD, Davey Smith G, Emmett PM, et al. FADS2 polymorphisms modify the effect of breastfeeding on child IQ. PLoS One. 2010;5:e11570.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Steer CD, Lattka E, Koletzko B, et al. Maternal fatty acids in pregnancy, FADS polymorphisms, and child intelligence quotient at 8 y of age. Am J Clin Nutr. 2013;98:1575–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Costea I, Mack DR, Lemaitre RN, et al. Interactions between the dietary polyunsaturated fatty acid ratio and genetic factors determine susceptibility to pediatric Crohn's disease. Gastroenterology. 2014;146:929–931.e3. [DOI] [PubMed] [Google Scholar]
- 80. Hovsepian S, Javanmard SH, Mansourian M, et al. Interaction of GCKR, MLXIPL and FADS genes polymorphisms with obesity in the occurrence of childhood metabolic syndrome. Middle East J Fam Med. 2018;16:20–28. [Google Scholar]
- 81. Sun C, Zou M, Wang X, et al. FADS1-FADS2 and ELOVL2 gene polymorphisms in susceptibility to autism spectrum disorders in Chinese children. BMC Psychiatry. 2018;18:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Brookes KJ, Chen W, Xu X, et al. Association of fatty acid desaturase genes with attention-deficit/hyperactivity disorder. Biol Psychiatry. 2006;60:1053–1061. [DOI] [PubMed] [Google Scholar]
- 83. Golding J, Northstone K, Emmett P, et al. Do omega-3 or other fatty acids influence the development of ‘growing pains’? A prebirth cohort study. BMJ Open. 2012;2:e001370.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Moltó-Puigmartí C, van Dongen MCJM, Dagnelie PC, et al. Maternal but not fetal FADS gene variants modify the association between maternal long-chain PUFA intake in pregnancy and birth weight. J Nutr. 2014;144:1430–1437. [DOI] [PubMed] [Google Scholar]
- 85. Bernard JY, Pan H, Aris IM, et al. Long-chain polyunsaturated fatty acids, gestation duration, and birth size: a Mendelian randomization study using fatty acid desaturase variants. Am J Clin Nutr. 2018;108:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu X, Wang G, Hong X, et al. Associations between gene polymorphisms in fatty acid metabolism pathway and preterm delivery in a US urban black population. Hum Genet. 2012;131:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Patterson E, Wall R, Fitzgerald GF, et al. Health implications of high dietary omega-6 polyunsaturated fatty acids. Ann Nutr Metab. 2012;2012:539426.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.