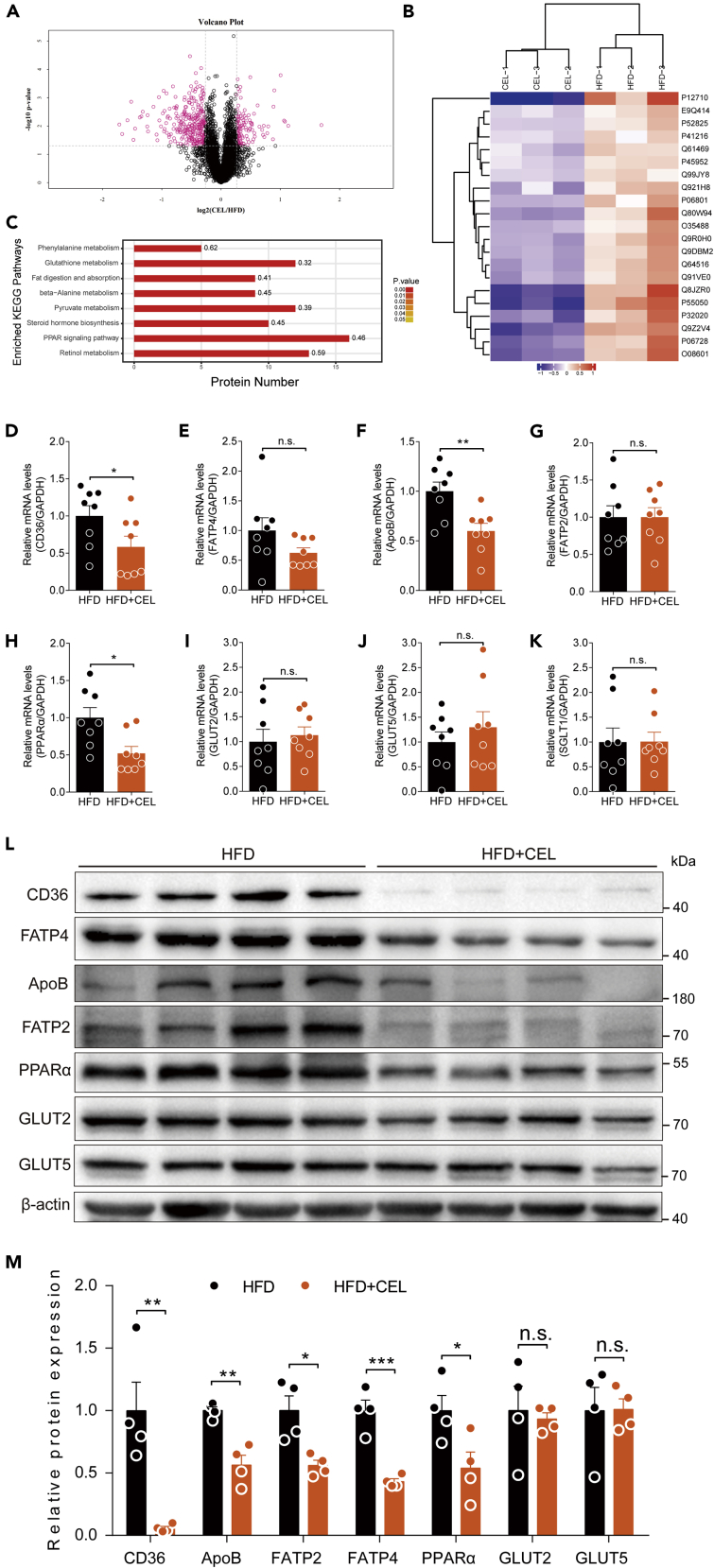
Figure 3.
Celastrol inhibits the expression of intestinal lipid transport-related genes in DIO C57BL/6 mice
(A–C) The intestinal protein of mice was identified and analyzed by proteomics. (A) The volcano plot is used to show significant differences between the two sets of sample data (n = 3 for each group). Red dots represent significantly differentially expressed proteins (fold changes greater than 1.2-fold and p value <0.05), with black dots representing proteins with no differential changes. (B) Heatmaps representing the selected 21 significantly differentially expressed proteins. Red represents significantly upregulated proteins, and blue represents significantly downregulated proteins. (C) KEGG enrichment was used to analyze the effect of celastrol on protein expression in the intestine of DIO C57BL/6 mice treated with celastrol or not (n = 3 for each group). The number on the right of the column on the histogram represents the enrichment factor. The enriched KEGG pathways with p < 0.05 are shown.
(D–K) mRNA levels of intestinal lipid and glucose-transport-related genes of mice treated with celastrol or not (n = 8 for each group). The genes included CD36 (D), FATP4 (E), ApoB (F), FATP2 (G), PPARα (H), GLUT2 (I), GLUT5 (J), and SGLT1 (K).
(L and M) (L) Lipid- and glucose-transport-related protein levels and (M) quantified signal intensity.
Error bars represent the mean ± SEM. p values were determined by Student's t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; n.s., not significant (p > 0.05).