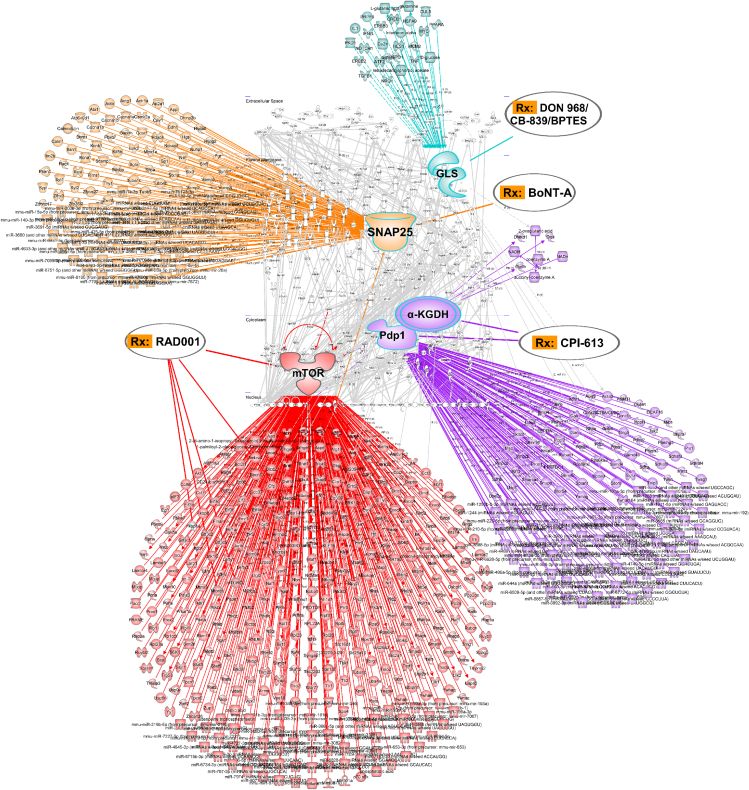
Figure 6.
Drug target prediction and repurposing
“Waterdrop” diagrams showing drug-target. Interaction prediction and computational drug repositioning from transcriptomics data in mouse GC created in Ingenuity Pathway Analysis (IPA). Note: nodes of RAD001-targeted mTOR (marked in red), CPI-613-targeted PDP1 and/or α-KGDH (purple), BoNT-A-targeted SNAP25 (yellow), and DON/968/CB-839/BPTES-targeted GLS (light blue). Lines represent biological relationships between molecules that include proteins, genes, mRNAs, microRNAs, and metabolites, generated from differentially expressed drug target genes (only drug targets differentially expressed between mouse GC versus WT at p < 0.05, q < 0.05 are shown, light gray). Molecule nodes are fetched from RNA sequencing data in mouse GC versus WT, edges are generated based on causal information in the Ingenuity Knowledge Base. The overlay-tool in IPA is used to predict drugs for indicated nodes.
See also Figure S3.