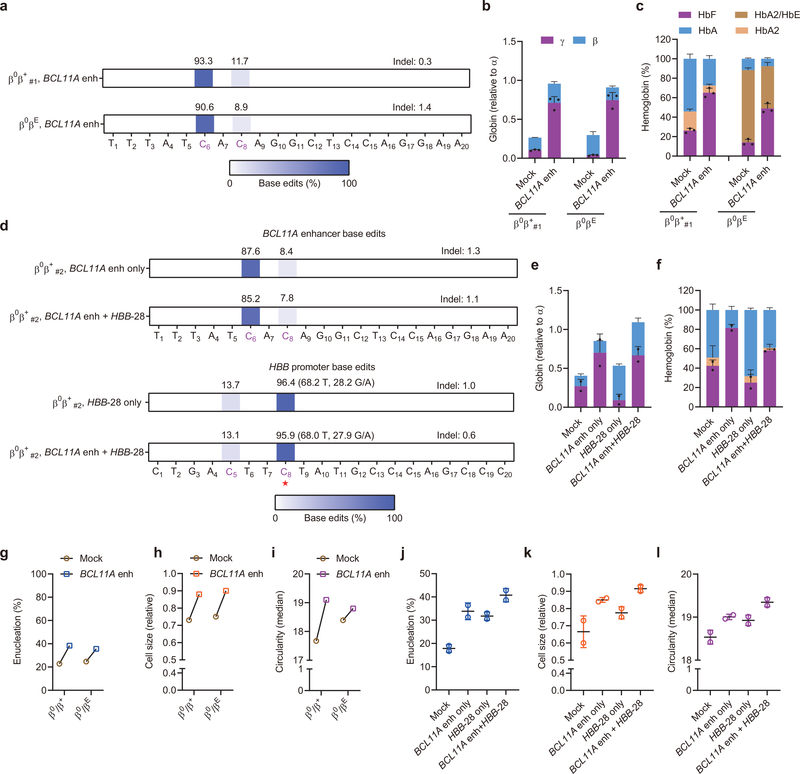
Fig. 3 |. Therapeutic and multiplex base editing in β-thalassemia patient CD34+ HSPCs.
a, Base editing in β-thalassemia donors β0β+ #1 and β0βE CD34+ HSPCs by A3A (N57Q)-BE3:sgRNA-1620. Two cycles of HSPC electroporation with 40 μM RNP were performed, separated by 24 hours. Base edits were measured by deep sequence analysis. b, β-like globin expression by RT-qPCR normalized by α-globin, measured from edited erythroid progeny. Data are plotted as mean±s.d. n=3 replicates from independent differentiation cultures. c, Hemoglobin levels by HPLC analysis. Data are plotted as mean±s.d. n=3 replicates from independent differentiation cultures. d, Base editing by A3A(N57Q)-BE3 at BCL11A +58 enhancer with sgRNA-1620 (top two rows) and HBB promoter by sgRNA-HBB-28 (bottom two rows) following single or multiplex editing in β-thalassemia donor β0β+#2 with HBB-28A>G β+ mutation. At the HBB-28A>G mutation position (noted with red asterisk, on opposite strand to spacer), alleles are divided into corrective C>T edits and alternative C>G/A edits. Because the donor is heterozygous at position C8, ~50% of alleles are T in unedited cells. e, β-like globin expression by RT-qPCR normalized by α-globin. Data are plotted as mean±s.d. n=2 replicates from independent electroporation. f, Hemoglobin levels by HPLC analysis. Data are plotted as mean±s.d. n=2 replicates from independent electroporations. g, j, Enucleation of in vitro differentiated erythroid cells from BCL11A enhancer only or multiplex editing experiments respectively. Data are plotted as mean±s.d. n=2 replicates from independent electroporations. h, k, Cell size by relative forward scatter intensity of enucleated erythroid cells, normalized to healthy donor, from BCL11A enhancer only or multiplex editing experiments respectively. Data are plotted as mean±s.d. n=2 replicates from independent electroporations. i, l, Circularity of enucleated erythroid cells by imaging flow cytometry, from BCL11A enhancer only or multiplex editing experiments respectively. Data are plotted as mean±s.d. n=2 replicates from independent electroporations.