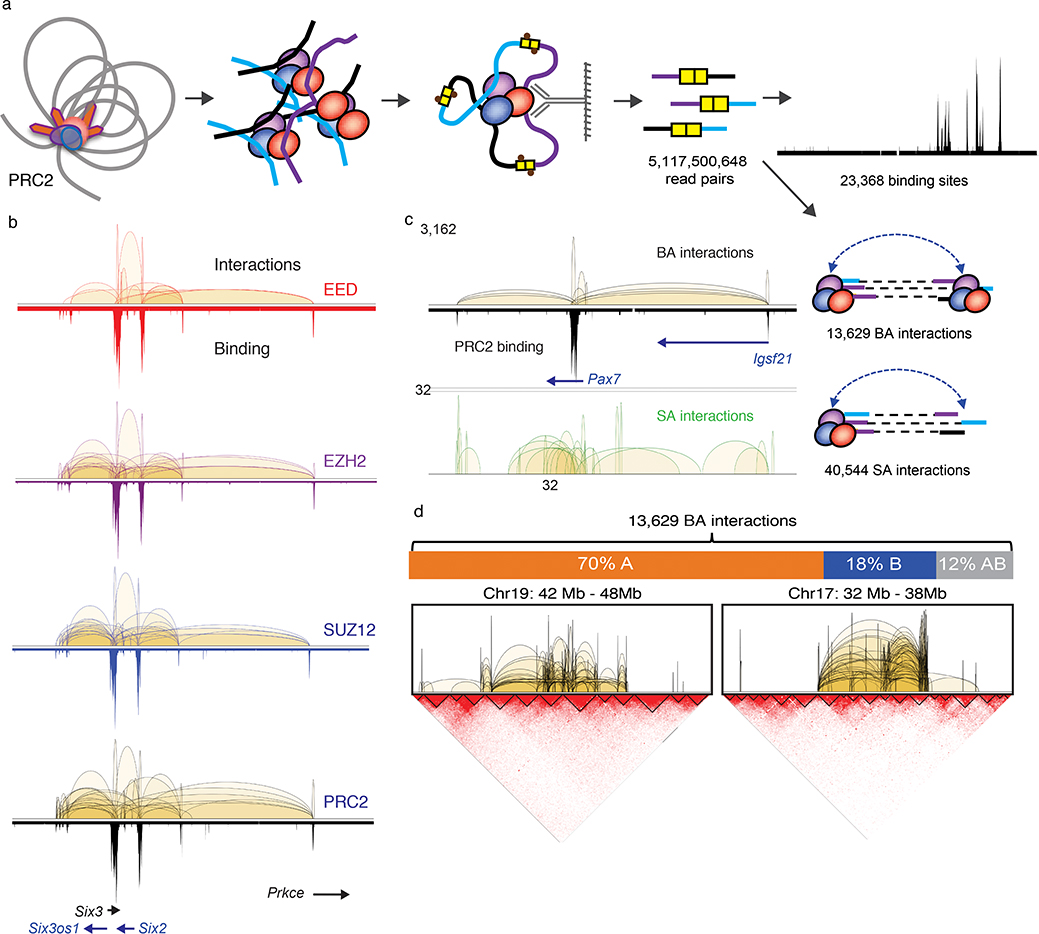
Fig. 1: ChIA-PET analysis defines PRC2 interactome in mESC.
a. Cross-linked chromatin was fragmented and subjected to proximity ligation followed by ChIP enrichment for three core PRC2 components, EED (n = 6), EZH2 (n = 7) and SUZ12 (n = 11) in mESC. See Supplementary Table 1 for sample details. Five billion read pairs were pooled to define PRC2 binding sites and interactions supported by PRC2 binding at both anchors (BA) and single anchor (SA). b. Interactions (upper tracks) and binding (lower tracks) profiles across chr17:85,366,518–86,405,710 region for EED (red), EZH2 (purple), SUZ12 (blue) and combined PRC2 (black) are displayed with matching gene track. c. BA and SA interactions across chr16:96,921,289–98,008,954 region are shown together with the PRC2 binding profile and the associated genes. Y-axis shows the interaction frequency represented by the number of PET counts. d. Upper Panel: The distribution of PRC2 BA interactions among nuclear compartments A, B and across A-B. Percent of total BA interactions are shown. Lower panel: ChIA-PET interactions within 6 Mb of chromosomes 17 and 19 are shown in reference with the topological associated domains (TADs) defined by Hi-C contact maps.