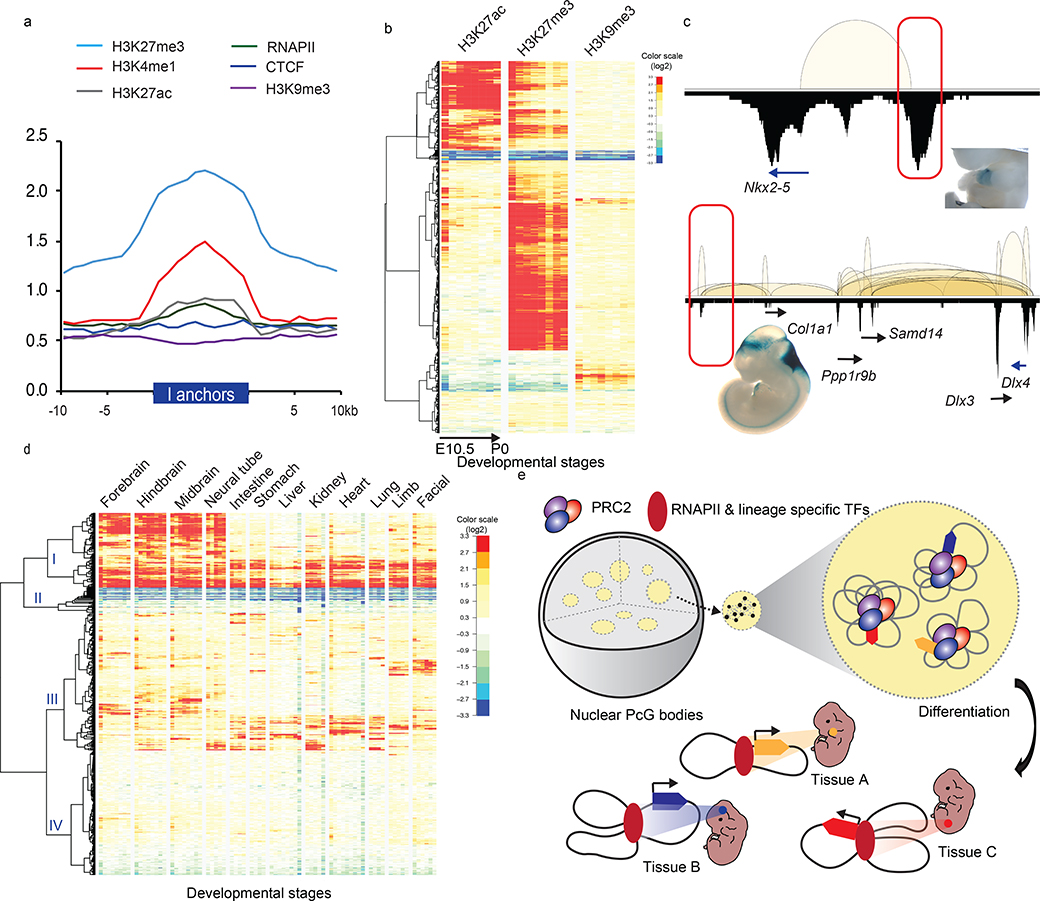
Fig. 5: Intergenic anchors exhibit the poised chromatin state and acquire enhancer signature during differentiation.
a. Enrichment fold of four histone modifications, RNAPII and CTCF binding over input control across ±10Kb of intergenic (I)-anchor regions. b. Heatmaps of H3K27ac, H3K27me3 and H3K9me3 normalized enrichment of the 1,800 I-anchors throughout progressive developmental stages in forebrain. The color scales represented the fold enrichment of ChIP over input.c. Enhancer activities of the PRC2 bound intergenic anchors in Nkx2–5 and Dlx3/4 loci observed in developing mouse embryos (heart in upper panel, mm1645 and hindbrain in lower panel, mm568) (www.enhancer.lbl.gov). d. Four distinct patterns of I-anchors based on the clustering of H3K27ac signal profiles across 74 different developmental stages collected from 12 tissues. The color scales represented the fold enrichment of ChIP over input. e. A model describes how PRC2 associated repressive chromatin foci contributing to TGS and transition into tissue specific enhancers during differentiation. PRC2 aggregated clusters are formed by extensive chromatin looping between silenced genes and their corresponding DREs. Upon differentiation, they are selectively dissolved, presumably in the absence of PRC2 binding. DREs acquire tissue specific enhancer signal and associate with RNAPII to activate their target gene expression.