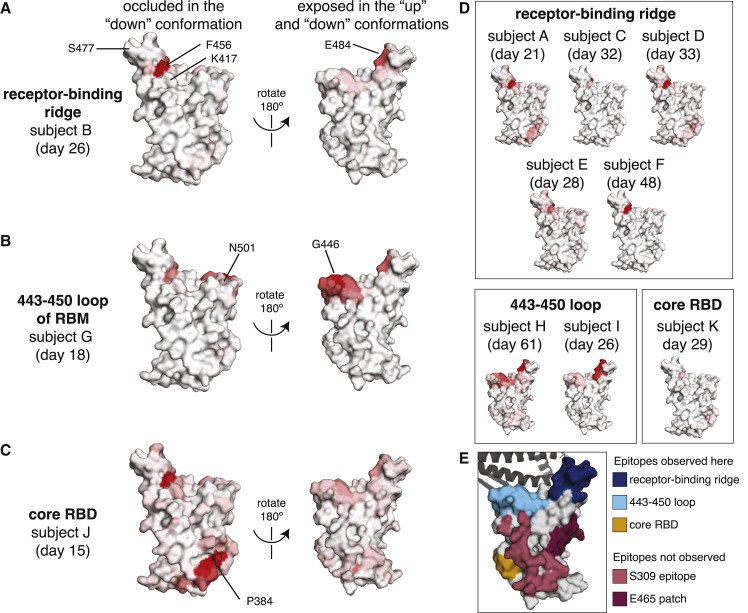
Figure 3.
Regions of the RBD where mutations strongly reduced binding by the antibodies in plasma collected from 11 individuals
The total effect of mutations at each site (sum of escape fractions) are projected onto the structure of the RBD (PDB: 6M0J), with white indicating no effect of mutations at that site and red indicating a large reduction in antibody binding. Two views of the RBD are shown: the surface of the RBD that is buried in the “down” conformation and the surface that is always exposed and accessible (Walls et al., 2020; Wrapp et al., 2020).
(A) For some individuals (typified by subject B), antibody binding is predominantly reduced by mutations in the receptor-binding ridge, particularly at sites F456 and E484.
(B) For some individuals (typified by subject G), antibody binding is strongly reduced by mutations in the 443–450 loop of the RBM in addition to the receptor-binding ridge.
(C) For a few individuals (typified by subject J), antibody binding is affected by mutations in the core RBD epitope around site P384.
(D) Samples from the other eight individuals fall in one of the three classes detailed in panels (A–C). For panels (A–D), the white-to-red coloring scale is set to span the same range as the y axis limits for that plasma in Figure 2.
(E) Mutations in two major surface regions (the S309 epitope and the sites near E465) do not strongly affect plasma antibody binding for any of the subjects. Shown is a surface representation of the RBD, with the three polyclonal plasma epitopes colored as in Figure 2. The S309 epitope and region near E465 (“E465 patch”) are shown in pink and maroon. ACE2 is shown in a dark gray cartoon representation.
Interactive versions of these structural visualizations are available at https://jbloomlab.github.io/SARS-CoV-2-RBD_MAP_HAARVI_sera/.