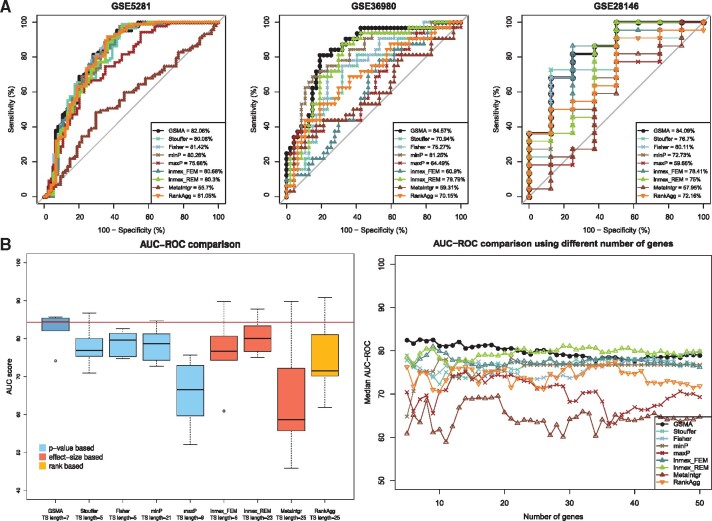
Fig. 3.
A comparison between the proposed meta-analysis framework—GSMA and eight other existing meta-analysis approaches—Stouffer’s method, Fisher’s method, minP, maxP, inmex_FEM, inmex_REM, MetaIntegrator and RankAggreg, using AD datasets. Panel A shows the AUC plots across three (out of six) independent validation datasets based on the test signature identified by each framework. For each of these three datasets, GSMA achieved higher AUC-ROC score compared to other approaches. The left plot in panel B shows the comparison of the AUC-ROC scores across all six validation datasets. The median AUC-ROC score obtained by using GSMA is significantly higher than the median AUC-ROC scores obtained by each category of approach(es) (P-value = 0.009 for four other P-value-based approaches, P-value = 0.045 for three other effect-size based approaches, P-value = 0.047 for the rank aggregation based approach, using Wilcoxon rank sum test). Finally, the right plot in panel B shows that, regardless of the length of the test signature, GSMA achieved higher average AUC-ROC scores compared to the others approaches in most of the cases