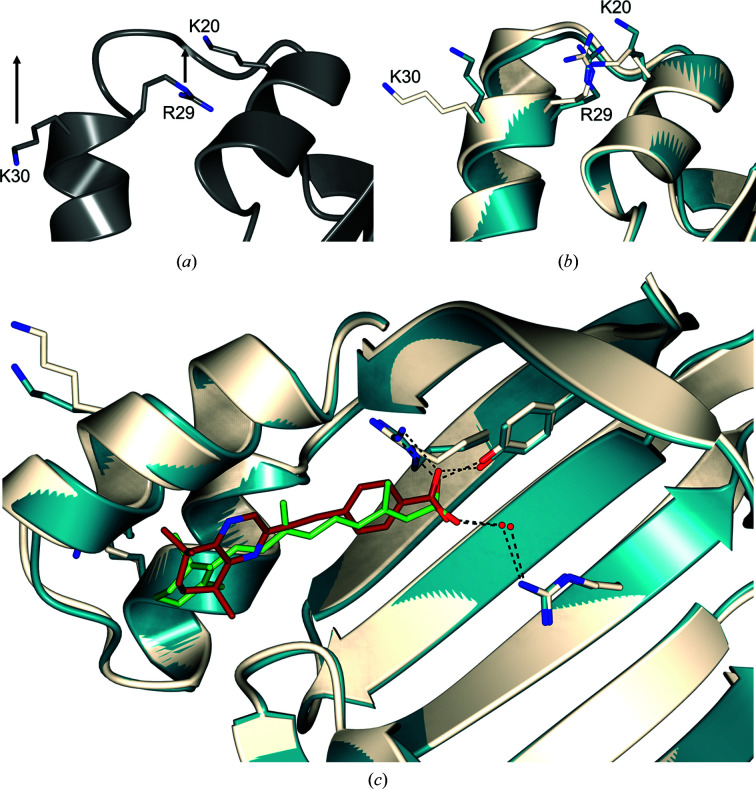
Figure 7.
Comparison of (a) unoccupied CRABPII (PDB entry 1xca, grey) and (b) the DC645 ligand-occupied complex (blue) aligned with the ATRA ligand-occupied structure (PDB entry 1cbs, white). Arrows denote the direction of movement as residues adopt the ligand-occupied conformation. (c) Cut-away alignment of CRABPII–DC645 (dark blue) and PDB entry 1cbs (white), showing the similarity between the orientation of DC645 (brick red; O atoms, red; N atoms, blue) and ATRA (green; O atoms, red; N atoms, blue) and their interaction with the binding triad Arg112-Arg133-Tyr135 and a conserved water molecule. The r.m.s.d. on Cα atoms is 0.48 Å.