Fig. 6.
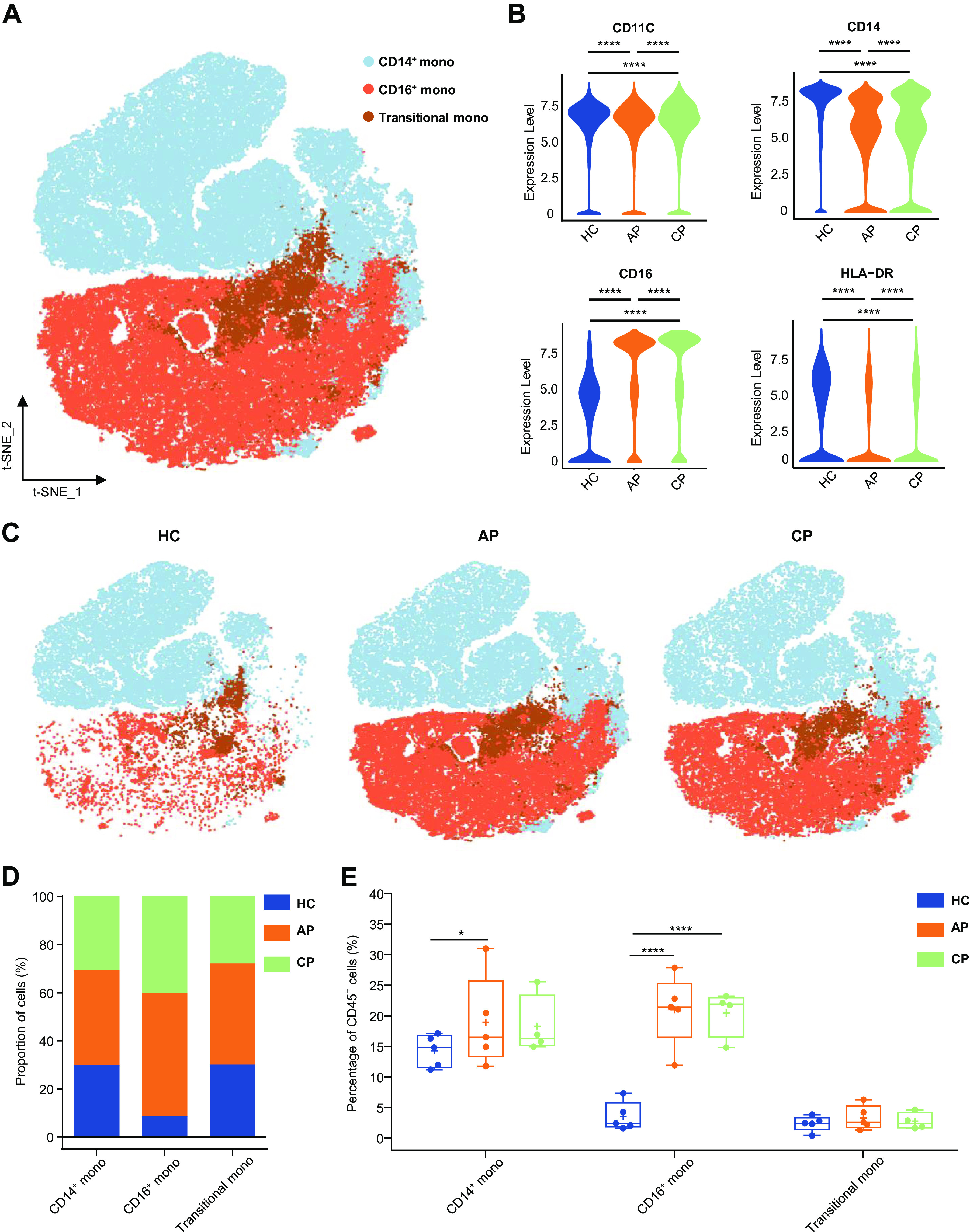
Characterization of single-cell monocytes from mass cytometry data. A: t-distributed stochastic neighbor embedding (t-SNE) plot of major monocyte subsets. Cells are colored on the basis of cell types. B: violin plot showing the expression of CD11C, CD14, CD16, HLA-DR in the monocytes cluster between (HC), acute phase (AP) and CP group. C: t-SNE projections of major monocyte subsets from the HC, AP, and CP group, respectively. D: bar plots highlighting cell abundances across monocyte subsets for the HC, AP, and CP group. E: percentage of major monocyte subsets in all blood CD45+ immune cells from the HC (n = 5), AP (n = 5), and CP group (n = 4). Adjusted P values are based on Wilcoxon rank-sum test (marker level) or two-way ANOVA tests with Bonferroni’s post hoc correction (parametric) between groups. ****Significant with adjusted P value < 0.0001. *Significant with adjusted P value < 0.05.
