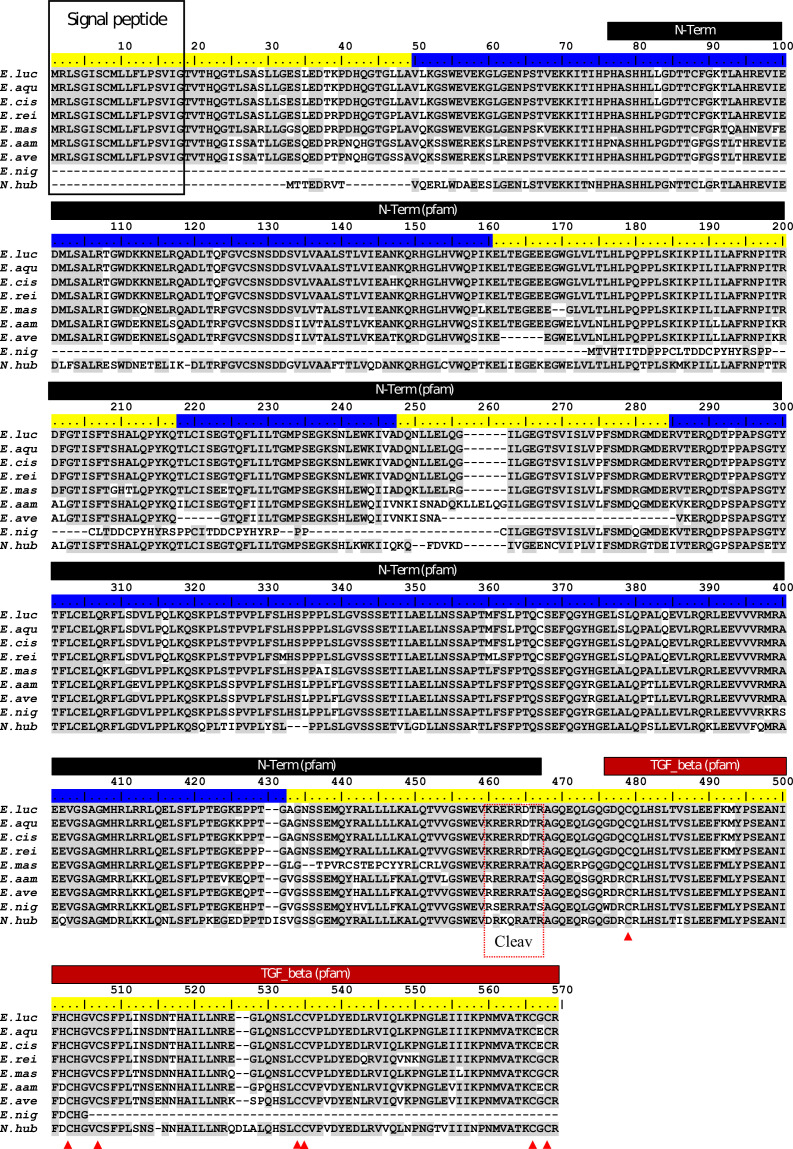
Appendix 1—figure 6. ClustalW alignment of Esocidae Amhby putative protein sequences.
The signal peptide was predicted with SignalP (Nielsen, 2017). No signal peptide was detected in the Amhby sequences of both E. niger and N. hubbsi. N-terminal region (indicated by a black bar) and the Transforming growth factor beta like domain (indicated by a red bar) were predicted with the Motif Scan tool at ExPASy (Gasteiger et al., 2003) with the Pfam motif database (Finn et al., 2014) optimized for local alignments (pfam-fs, Pfam 32.0, September 2018). The seven exons of amhby are shown by the alternating blue and yellow colors on the sequence ruler. The seven conserved cysteines of the TGF-beta domain are indicated by red arrowheads. The region containing the putative Amh cleavage site (Cleav) is boxed in red. E. luc (Esox lucius), E. aqu (E. aquitanicus), E. cis (E. cisalpinus), E. rei (E. reichertii), E. mas (E. masquinongy), E. aam (E. americanus americanus), E. ave (E. americanus vermiculatus), E. nig (E. niger) and N. hub (Novumbra hubbsi).