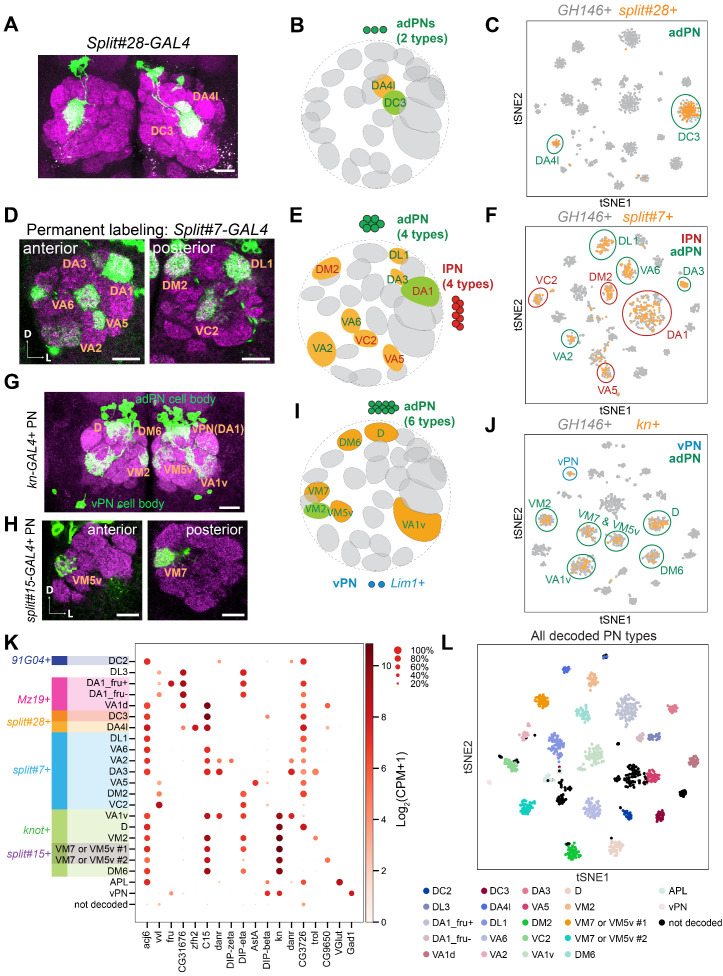
Figure 2. Matching 15 transcriptomic clusters to specific projection neuron (PN) types at 24 hr APF.
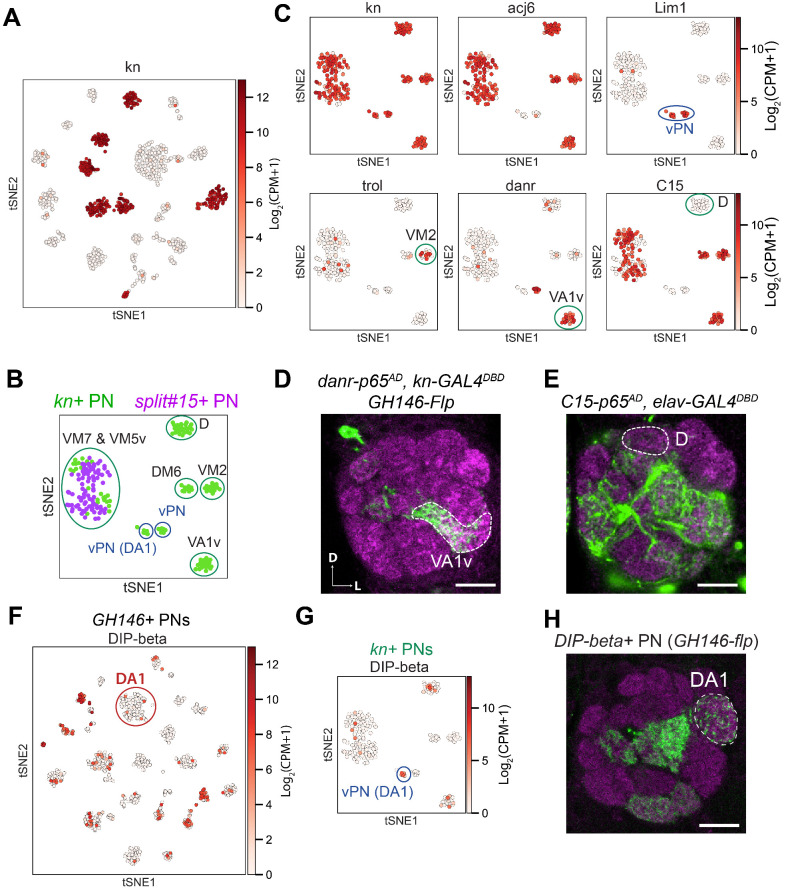
(A) Representative maximum z-projection of confocal stacks of split#28 GAL4 in adults. Dendrites of split#28 GAL4+ PNs target the DC3 and DA4l glomeruli. (B) Diagram of split#28-GAL4+ PNs. (C) tSNE plot showing newly sequenced split#28-GAL4+ PNs, which form two clusters that can be assigned to DC3 and DA4l PNs (see also Figure 2—figure supplement 1). (D) Representative confocal images of split#7 GAL4 labeled PNs using permanent labeling strategy. One anterior section and one posterior section of the antennal lobe are shown. Using permanent labeling, we found that this driver is expressed in eight PN types. Genotype: split#7-GAL4, UAS-Flp, Actin promoter-FRT-STOP-FRT-GAL4, UAS-mCD8-GFP. (E) Diagram of split#7-GAL4+ PNs. split#7 GAL4 labels eight types of PNs. four from the adPN lineage (green letters) and four from the lPN lineage (red letters). (F) tSNE plot of split#7 GAL4 PNs with GH146+ PNs (see Figure 2—figure supplement 2 for details on the decoding procedure). (G) Representative maximum z-projection of confocal stacks of kn+ PNs in the adult. kn-GAL4 was intersected with GH146-Flp to restrict the expression of GAL4 in only PNs. (H) Representative confocal images of split#15 GAL4 in adults, which labels two kn+ PN types. (I) Diagram showing that kn+ PNs include six types of adPNs and two vPNs. (J) tSNE plot of kn-GAL4 PNs with GH146+ PNs (see Figure 2—figure supplement 3 for details on the decoding procedure). (K) Dot plot summarizing drivers and marker genes we used to map 21 transcriptomic clusters to 20 PN types [14 adPNs, 5 lPNs—DA1 PNs form two clusters, one fru+ and one fru– (Li et al., 2017)—and 1 vPNs] and the anterior paired lateral (APL) neurons at 24 hr APF. Gene expression level [log2(CPM+1)] is shown by the dot color, and percentages of cells expressing a marker are shown by dot size. (L) tSNE plot showing 24 hr APF PNs colored by PN types (GH146+ PNs with split#7+/ split#28 PNs to increase cell number in some less abundant PN types). Scale bars, 20 μm. Axes, D (dorsal), L (lateral). In panel B, E, and I, orange glomeruli represent PN types of unknown transcriptomic identity prior to this study. Green glomeruli represent PN types whose transcriptomic identity were previously decoded. Note that the positions of cells on a tSNE plot are dependent on the random initialization of the program as well as every cell present in the dataset, therefore the position of GH146+ PNs clusters are different when we plot them with different set of newly sequenced PNs (gray in panels C, F, and J).