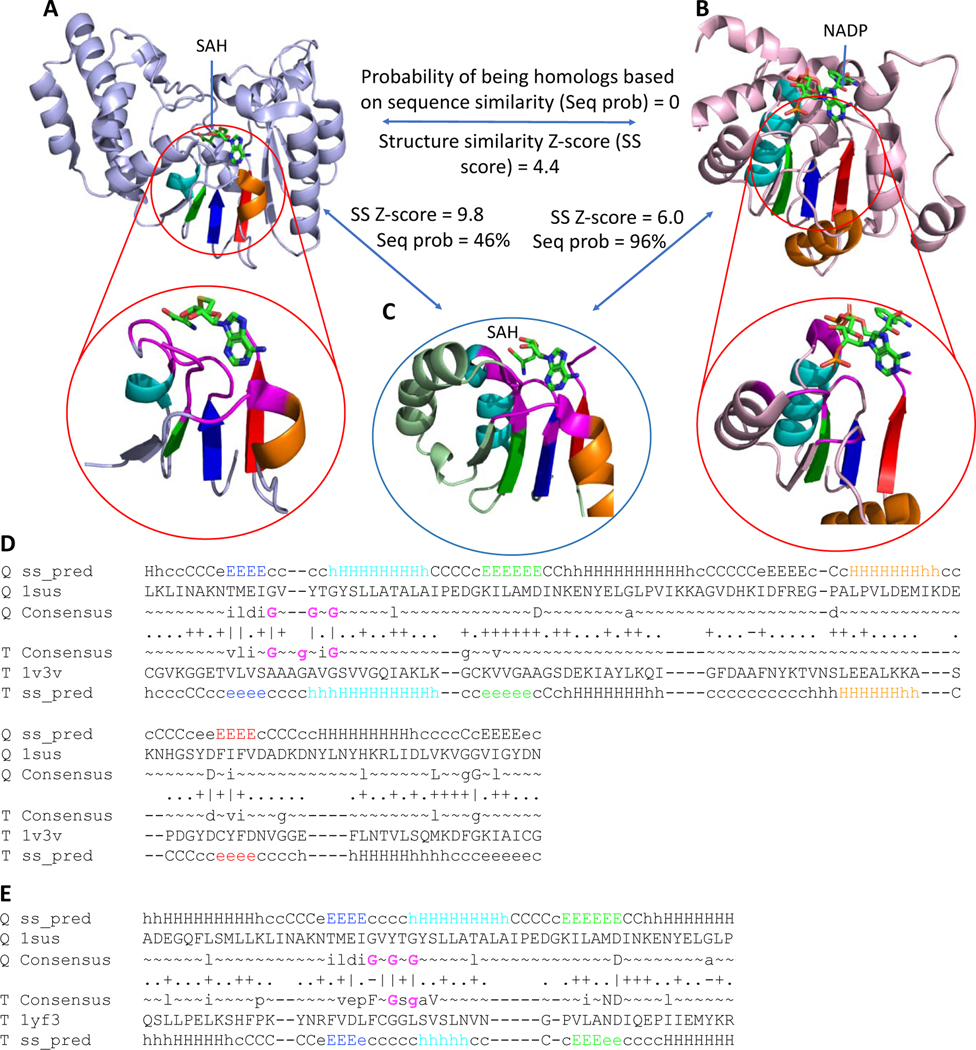
Fig 4. Example of distant homology that can be detected by active site similarity.
RLM SSEs are colored in rainbow with active site residues shown in magenta. (A) DNA-adenine methyltransferase (PDB: 1yf3) binds SAH. (B) 12-hydroxydehydrogenase /15-Oxo-prostaglandin 13-reductase (PDB: 1v3v) binds NADP. (C) Caffeoyl-CoA O-methyltransferase (PDB: 1sus) binds SAH. Sequence homology probability was calculated using HHpred [49], structure similarity score was calculated using DALI [59]. (D) HHpred sequence alignment of caffeoyl-CoA O-methyltransferase (PDB: 1sus) and 12-hydroxydehydrogenase / 15-Oxo-prostaglandin 13-reductase (PDB: 1v3v). Gly residues from Gly-rich loop are colored by magenta. (E) HHpred sequence alignment of caffeoyl-CoA O-methyltransferase (PDB: 1sus) and DNA-adenine methyltransferase (PDB: 1yf3). Gly residues from Gly-rich loop are colored by magenta.