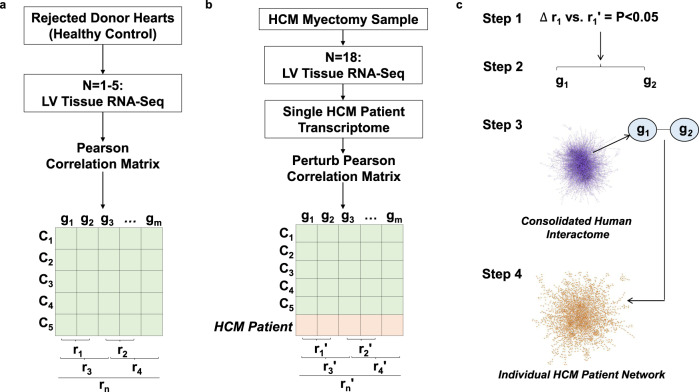
Fig. 1. Strategy for developing patient-specific protein–protein interaction networks in HCM.
a Left ventricular (LV) myocardial samples from rejected heart transplant donors serving as healthy controls (N = 5) (C1–C5) were analyzed using RNA-Seq, and the Pearson correlation coefficient (r) was calculated for all gene (g) pairs. n, total combination of pairwise correlations; m, total number of genes. b Anterior septal myectomy samples from hypertrophic cardiomyopathy (HCM) patients were analyzed using RNA-Seq. The transcriptomic profile of an individual HCM patient was added to the control gene expression matrix, and the new Pearson correlation coefficient (r′) was calculated for all gene pairs. The HCM patient transcriptomic profile was then removed from the matrix and the process was repeated sequentially for the HCM (N = 18). c (Step1) Statistically significant changes between the r and r′ coefficients were collected, and (Step 2) those gene pairs (g1–g2) were mapped to the consolidated human interactome (Step 3), which contains information on 15,489 proteins and 188,973 protein–protein interactions (PPIs). The statistical test used in this step was the two-tailed Z-test, and the P values were adjusted by the Bonferroni correction procedure for multiple comparison. In Step 4, gene pairs for which a PPI was identified in the consolidated human interactome were used to generate individual-patient PPI network reticulotypes.