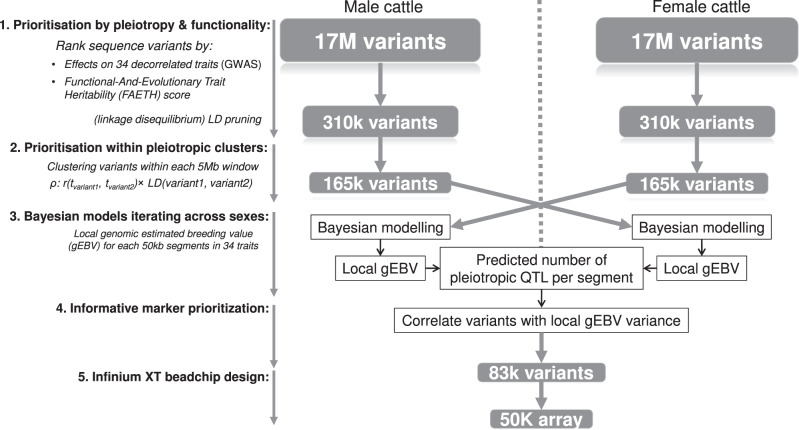
Fig. 1. Overview of the 5-step genome-wide fine-mapping analysis with 34 traits.
Steps 1–2 that prioritised variants based on the multi-trait GWAS and functional information were conducted separately in the male and female cattle. The design of the Bayesian mixture modelling across two sexes (step 3) aimed at avoiding the bias of variant preselection and is detailed in Supplementary Note 1. Variant effects from Bayesian models were used to calculate local gEBV for each 50 kb segment. Different sets of local gEBV allowed the estimation of the predicted number of pleiotropic QTL per segment. Within each segment, variants with the highest correlation with the local gEBV variance were selected (step 4). These variants were used to customise the XT-50K bovine genotyping array (step 5).