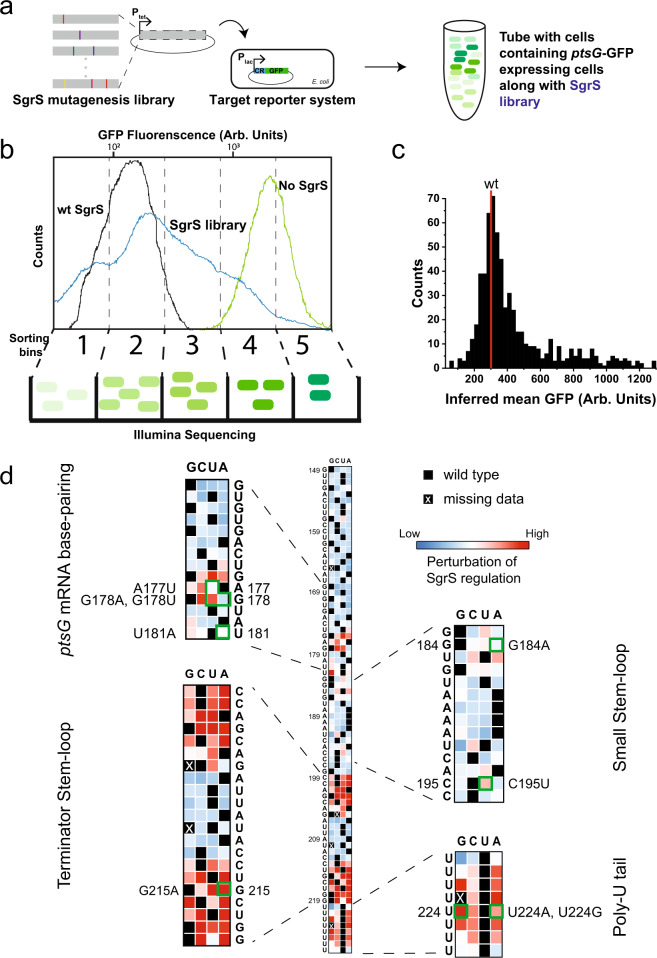
Fig. 2. Mapping efficacy of SgrS regulation of ptsG mRNA with respect to its sequence.
a Preparation of SgrS mutation library. Mutations were introduced into the SgrS plasmid using mutagenesis PCR. The mutations introduced are represented by the colored bars. This library was then transformed into an E. coli strain already transformed with the ptsG mRNA plasmid fused with a GFP reporter. The varying levels of GFP fluorescence are shown by the green colors. b Sorting of the cells and sequencing. The cells with two-plasmid co-expression were sorted using flow cytometry. The SgrS library (blue) shows GFP fluorescence that spans the region from the wild-type SgrS (black) to target (ptsG)-only (green). Cells were sorted into five evenly spaced (log scale) fluorescence bins and the occupancy percentages were 18.74%, 33.76%, 30.91%, 13.83%, and 2.76%, respectively. The cells from each bin were grown, DNA was purified, barcoded, and sequenced using Illumina sequencing platform. c Histogram of the Sort-Seq measurements from the SgrS library from two replicates combined. The mean fluorescence for the wild-type SgrS is shown in red. d Heatmap showing the effect of mutations on SgrS regulation of ptsG mRNA starting from nucleotide 149 to 227. The colors in the boxes are scaled from blue (low) to red (high), according to the level of perturbation of SgrS regulation. Black squares represent the wild-type base at each position and the black boxes with white crosses show the positions of the mutants missing in the experiment. Text shows the wild-type sequence of SgrS. Insets show the four regions of SgrS, viz. base-pairing region, small stem-loop, terminator stem-loop, and the poly-U tail. Source data are provided as a Source data file.