Figure 5. PDPK1 is a high-affinity WIN site binding protein.
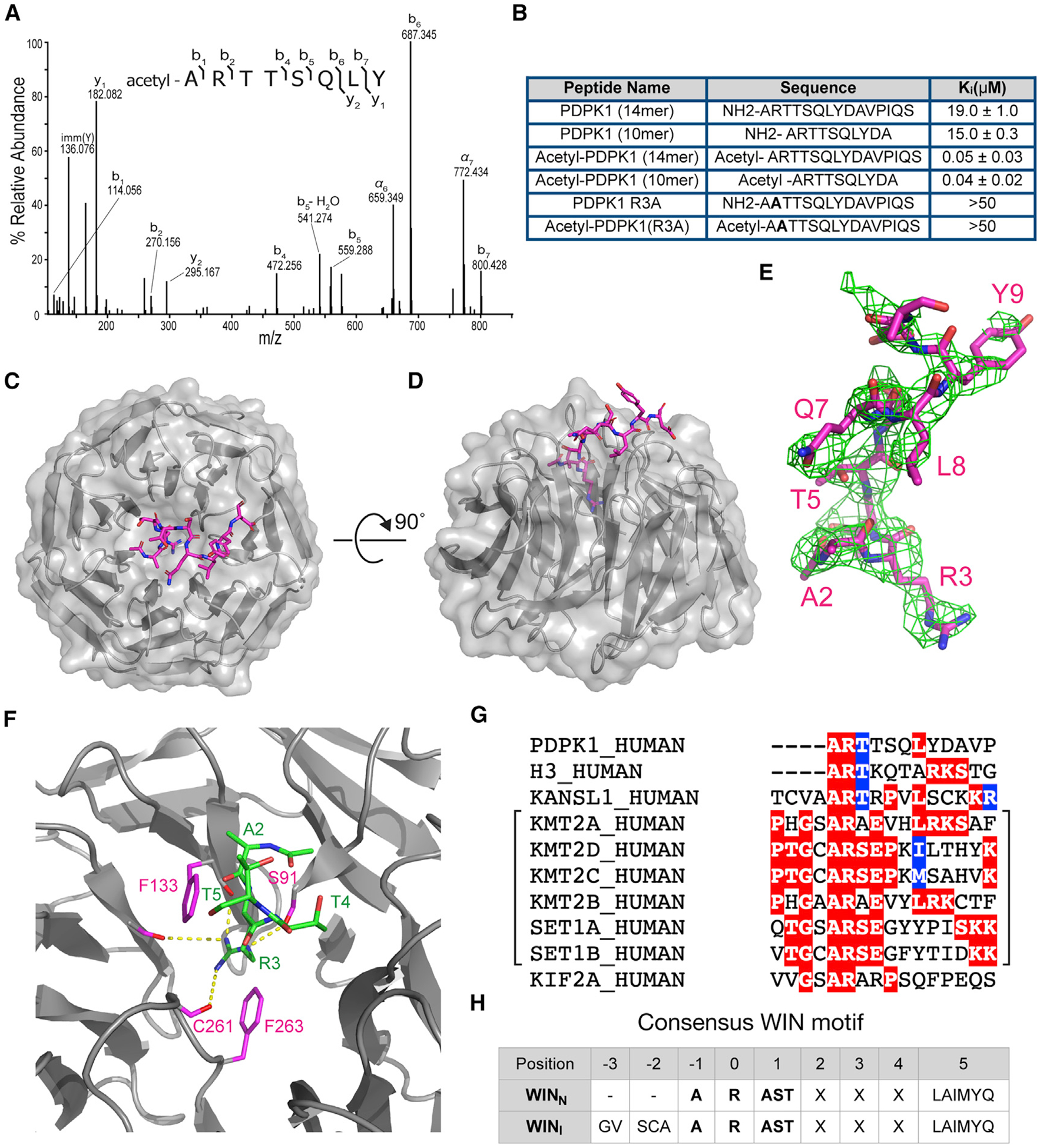
(A) Tandem mass spectrum of N-terminally acetylated PDPK1 peptide, residues 2–9. The doubly protonated precursor, [M+2H]+2, with m/z 491.2556 was fragmented with higher-energy collisional dissociation. The identified amino acid sequence is provided above the annotated spectrum; brackets indicate sites of dissociation at the peptide backbone. Observed product ions are assigned to their corresponding m/z peaks in the mass spectrum.
(B) Binding constants of PDPK1 peptides were determined using a TR-FRET-based KMT2A peptide competition assay. All peptides are amidated at the C terminus. Two or more repeats were obtained; average Ki values and standard deviations are reported.
(C) Structure of WDR5 in complex with the acetylated-PDPK1 WIN peptide. The PDPK1 WIN peptide is shown in stick representation (magenta, colored by atom type); WDR5 is shown as cartoon with semitransparent surface representation (gray). 2.7Å resolution.
(D) As in (C) but rotated along a 90°axis.
(E) The Fo-Fc omit map of PDPK1 peptide bound with WDR5 domain contoured at 2.0 σ level. PDPK1 peptide is shown in magenta sticks.
(F) Close-up of the first three residues of PDPK1 (ART) in the WIN site of WDR5. The PDPK1 peptide is green sticks; WDR5 is gray ribbons. Key WDR5 residues F133, S175, C261, and F263 are indicated in pink stick representation. Yellow dotted lines indicate intermolecular hydrogen bonds.
(G) WIN motif of PDPK1 aligned with established WIN motifs. The related histone methyltransferase enzymes are grouped with brackets.
(H) Table summarizing consensus sequences for internal (WINI) and N-terminal (WINN) WIN motifs.
