Abstract
Background & Aims:
Methylation of lysines on histones, controlled by various methyltransferases and demethylases, is an important component of epigenetic modifications, and abnormal regulation of such enzymes serves as common events in hepatocellular carcinoma. We determined to identify important methyltransferases and demethylases that might regulate the development of hepatocellular carcinoma by bioinformatics.
Methods:
The Oncomine and UALCAN databases were used to retrieve mRNA expression levels of histone lysine methyltransferases and demethylases in hepatocellular carcinoma. Data analyses of genetic alterations, mainly mutations and copy number alterations, were performed on the cBioportal platform. Protein-protein interactions were established in the STRING database.
Results:
mRNA expression of 8 genes correlated with clinical staging and grading, whereas 4 genes indicated a role in the prognosis, all co-expressed with SEDB1 and WHSC1. Genetically, 12 genes showing an alteration rate higher than 5% were identified, and only 3 were indicative of prognosis. Copy number gains in ASH1L, SETDB1, and KDM5B might partially contribute to the upregulation of their mRNA expression. The close relationship of mutations in MLL2/MLL3 with driver gene mutations in hepatocellular carcinoma provided a rationale for further investigation.
Conclusions:
We identified 11 methyltransferases and demethylases for major histone lysines that might be promising research targets in the pathogenesis, development, and prediction of prognosis in hepatocellular carcinoma using bioinformatics.
Keywords: hepatocellular carcinoma, epigenetics, histone lysine, methyltransferases, demethylases, bioinformatics
Introduction
In contrary to a more stable overall cancer incidence rate, liver cancer has shown the largest increase in the incidence of 2.5% between 2012-2016 in America.1 Additionally, hepatocellular carcinoma (HCC), which mostly occurs in patients with underlying liver conditions, such as cirrhosis caused by hepatitis B or C virus,2,3 takes up the largest proportion of all primary liver cancers. As the third leading cause of cancer death worldwide, HCC has led to 746, 000 deaths solely in 2012.2 At present, the treatment of liver cancer includes liver transplantation, surgical resection, embolization, stereotactic radiotherapy, ablation, and chemotherapy.3,4 However, patients with HCC are usually at more advanced stages at first diagnosis when few therapeutic approaches are effective.5 And even though surgical resection may serve as a promising option for a small group of HCC patients, tumor recurrence deteriorates the condition in around 70% of these cases at 5 years.6,7
Understanding the pathogenesis of HCC is essential in determining the corresponding therapeutic targets for future investigation. Development of HCC has been regarded as a multistep process, where the accumulation of somatic DNA alterations and epigenetic modifications (especially DNA methylation and histone modifications) induced by chronic inflammation contributes to the heterogeneity of its molecular mechanisms.2,3 In various tumor models, including HCC, epigenetic modifications of tumor suppressor genes lead to the constitutive expression of oncogenes during the early stages of oncogenesis, and DNA alterations of the same oncogenes facilitate this process to accelerate tumor progression.8 Unlike DNA alterations, epigenetic modifications are characterized by heritable, reversible, and dynamic regulation of gene expression, thus drawing enormous attention from worldwide research centers in terms of therapeutic potential in recent years.
Methylation of lysines on histones is one of the crucial components in epigenetic modifications and has become a promising therapeutic target for various diseases, including cancer.9-12 Primary sites for lysine methylation in human beings are identified on histone H3 at lysine 4 (H3K4), lysine 9 (H3K9), lysine 27 (H3K27), lysine 36 (H3K36), and lysine 79 (H3K79), and on histone H4 at lysine 20 (H4K20).10 With the first discovery of histone lysine methyltransferase (KMT), SUV39H1, in 2000, the hunt for more histone KMTs and histone lysine demethylases (KDMs) kicked off.13 Current validated histone KMTs and KDMs of primary histone lysines were summarized in Table 1 (data from the GeneCards database, https://www.genecards.org/). Associations among methylation of histone lysine, regulation of gene expression, and oncogenesis offer a perspective that alterations in histone KMTs/KDMs impact the initiation and/or development of various cancer types.10 For example, in lung adenocarcinoma, the high mutation frequency of SETD2, a histone KMT of H3K36, serves as a reminder of the classical roles of other tumor suppressors.14
Table 1.
Major Methyltransferases and Demethylases of Lysines in Histone 3 and 4.
| H3K4 | H3K9 | H3K27 | H3K36 | H3K79 | H4K20 | |
|---|---|---|---|---|---|---|
| Methyltransferases | MLL1 (KMT2A) MLL2 (KMT2B) MLL3 (KMT2C) MLL4 (KMT2D) MLL5 (KMT2E) SET1A (KMT2F) SET1B (KMT2G) SETD7 (KMT7) SMYD1 (KMT3D) SMYD2 (KMT3C) SMYD3 (KMT3E) ASH1L (KMT2H) SETD3 SETMAR |
SETDB1 (KMT1E) SETDB2 (KMT1F) SUV39H1 (KMT1A) SUV39H2 (KMT1B) EHMT1 (KMT1D) EHMT2 (KMT1C) PRDM2 (KMT8) |
EZH2 (KMT6) | SETD2 (KMT3A) NSD1 (KMT3B) WHSC1 (NSD2) WHSC1L1 (NSD3) ASH1L (KMT2H) SETD3 SETMAR |
DOTH (KMT4) | SETD8 (KMT5A) SUV420H1(KMT5B) SUV420H2 (KMT5C) NSD1 (KMT3B) |
| Demethylases | KDM1A (LSD1) KDM1B (LSD2) KDM2B (JHDM1B) KDM5A (JARID1A) KDM5B (JARID1B) KDM5C (JARID1C) KDM5D (JARID1D) C14orf169 (RIOX1) |
KDM1A (LSD1) KDM3A (JHDM2A) KDM3B (JHDM2B) KDM4A (JHDM3A) KDM4B (JMJD2B) KDM4C (JMJD2C) KDM4D (JMJD2D) KDM7A (JHDM1D) PHF8 (KDM7B) |
KDM6A (UTX) KDM6B (JMJD3) PHF8 (KDM7B) KDM7A (JHDM1D) |
KDM2A (JHDM1A) KDM2B (JHDM1B) KDM4A (JHDM3A) KDM4C (JMJD2C) C14orf169 (RIOX1) |
/ | KDM7A (JHDM1D) PHF8 (KDM7B) |
Previous reports have supported the idea that aberrant regulation of histone KMTs and KDMs is a common event in HCC. For instance, over-expression of EZH2, the only known H3K27 methyltransferase, was common in human HCC tissues, and transcriptional silencing of CXCR4 by EZH2-mediated H3K27me3 was correlated with a poor prognosis in HCC patients.15 Copy number gain of SETDB1, a histone H3K9 methyltransferase, was reported to regulate the growth of HCC cells through methylation of a well-known tumor-suppressor p53.16 Nevertheless, the role of distinct histone KMTs and KDMs in the pathogenesis and development of HCC remains unorganized. With the current rapid construction of Bioinformatics, a comprehensive study of distinct histone KMTs and KDMs in HCC can systemically summarize and help reveal the underlying mechanisms in the pathogenesis of HCC and provide promising prognostic and therapeutic markers for future investigation.
In the present study, we addressed this topic by analyzing the mRNA expressions and genetic alterations of 49 histone KMTs and KDMs and their correlations with multiple clinical parameters in HCC patients using data from multiple databases. Additionally, we aimed to establish associations between these KMTs/KDMs and known crucial genes in HCC.
Materials and Methods
One of the core functions of the ONCOMINE platform (www.oncomine.org) is the differential analysis of gene expression in various cancer types, including liver cancer. The mRNA expression levels of 49 major methyltransferases and demethylases between multiple types of cancer tissues vs. adjacent normal tissues, and the number of significant unique analyses were retrieved from the platform, using the following criteria: P-value < 0.01; |Fold change| > 1.5; Gene rank, top 10%; Data type: mRNA. Genes showing only the over-expression profile or the low-expression profile in more than one significant unique analyses were selected, while genes with contradictory expression profiles or no significant unique analyses were excluded. Then we requested detailed information on the retrieved results in liver cancer to identify analyses performed between HCC tissues and normal lung tissues.
Retrieved methyltransferases and demethylases with altered mRNA expression were validated in the UALCAN database (http://ualcan.path.uab.edu). Genes with non-significant or contradictory results were excluded, while consistent results were collected and used for further analysis. Meanwhile, UALCAN also provides data concerning the association between the mRNA expression level and individual cancer stages/tumor grades/overall survival in patients with HCC. These associations were retrieved to help explain the potential roles that specific genes play in the diagnosis and prognosis of patients with HCC.
Co-expression analysis of the RNA-RNA interactions of methyltransferases/ demethylases for histone lysine methylation in HCC was performed through the ENCORI platform (The Encyclopedia of RNA Interactomes, http://starbase.sysu.edu.cn). Results were extracted and visualized as a co-expression network. Associations with the correlation coefficient (|r|) > 0.5 and p < 0.05 were considered as significant co-expression between 2 genes and presented in the network.
Six studies involving HCC sample tissues in the cBioPortal (www.cbioportal.org) database were selected for further analyses: Hepatocellular Carcinoma (MSK, Clin Cancer Res 2018); Hepatocellular Carcinomas (INSERM, Nat Genet 2015); Liver Hepatocellular Carcinoma (AMC, Hepatology 2014); Liver Hepatocellular Carcinoma (RIKEN, Nat Genet 2012); Liver Hepatocellular Carcinoma (TCGA, Firehose Legacy); Liver Hepatocellular Adenoma and Carcinomas (MSK, PLOS One 2018). A total of 1,085 patients and 1,089 samples were obtained from the above studies. Results concerning mutations and putative copy-number alterations of 49 major methyltransferases and demethylases were retrieved by cBioPortal (www.cbioportal.org) in sample tissues under the following criteria: 1) cancer type: hepatocellular carcinoma; 2) number of samples per patient: 1; 3) overall survival status: living or deceased; 4) with mutation data: yes; 5) with copy number alteration (CNA) data: yes. After screening with our criteria, only 353 samples were collected. And all these samples came from the Liver Hepatocellular Carcinoma (TCGA, Firehose Legacy). Meanwhile, we identified the top altered candidate driver cancer genes in the aforementioned sample tissues with the following criteria: 1) alteration frequency > 10%; 2) candidate driver cancer genes determined by the MutSig or the GISTIC algorithm. Mutual exclusivity or co-occurrence analyses among altered methyltransferases/ demethylases (mutations/CNAs) and the altered candidate driver cancer genes in HCC were performed as well. Alterations in selected methyltransferases/demethylases and their correlations with overall survival and disease/progression-free survival in HCC patients were demonstrated by Kaplan-Meier plotting.
To understand the possible mechanisms and associations of important methyltransferases and demethylases at the protein level in HCC, we constructed a PPI network. Known crucial genes that contribute to the development of HCC were obtained using the Phenolyzer software (http://phenolyzer.wglab.org) and were referred to as seed genes, using “hepatocellular carcinoma” as the disease term. Top 50 seed genes (protein-coding genes) were retrieved according to the rank of the score. Seed genes and identified methyltransferases/demethylases (associated with clinical features of HCC) were then entered into the STRING database (https://string-db.org/). Results were retrieved after choosing Homo sapiens as the organism and the minimum required interaction score as high confidence (0.700) and visualized in a PPI network. Disconnected nodes were not shown in the network.
All data are available on online platforms and databases. Parameters used within online platforms defaulted unless specifically indicated. Students’ t-tests were used to compare the difference of transcription expression in the Oncomine platform. The significance of difference in expression values retrieved from the UALCAN database was estimated by the student’s t-test considering unequal variance. P values between each group pair within all survival analyses were calculated by the log-rank test. P < 0.05 was considered statistically significant unless otherwise indicated.
Results
Abnormal mRNA Expression of Multiple Methyltransferases/Demethylases in HCC
The Oncomine platform was employed to understand the general mRNA expression levels of genes across various cancer types, especially in liver cancer. As was shown in Figure 1, the mRNA expression profiles of 49 methyltransferases/demethylases in 20 types of cancers, compared with those in normal tissues, were displayed. Significant up-regulation of 16 genes and down-regulation of 6 genes were identified in multiple analyses of liver cancer tissues vs. normal tissues (Table S1). Next, we explored the detailed significant analyses concerning these 22 genes to collect comparisons performed between HCC tissues and normal liver tissues. Among the 16 up-regulated genes, 13 of them were found to be included in at least 1 dataset comparing HCC and normal tissues, and 4 out of 6 down-regulated genes were collected as well (Table S1. In the Wurmbach liver dataset, EZH2 was shown with the biggest |fold change| of 5.261 (p = 9.41E-07), while KDM6B had the smallest |fold change| of 1.514 (p = 9.02E-05) in the Roessler liver dataset. Furthermore, we validated our results by UALCAN, which integrates enormous resources from the TCGA database. We found the results of all 13 up-regulated genes in the Oncomine platform were consistent with those in UALCAN, but only 1 out of 4 down-regulated genes, KDM6B, was observed to have a lower expression in HCC tissues by UALCAN (Figure 2A). Increasing mRNA expressions of MLL and KDM4B in HCC by UALCAN were contrary to our findings in the Oncomine platform, while no significant change was identified in the expression of KDM7A (Fig. S1). Therefore, these 3 genes were excluded from further analysis to increase data credibility. Together, we identified 14 methyltransferases/demethylases with abnormal mRNA expression in HCC using the Oncomine platform and the UALCAN database.
Figure 1.

Changes of transcription level among 49 major methyltransferases and demethylases between various types of cancer tissues and normal tissues (ONCOMINE). For each entry, mRNA expressions were displayed in the form of cancer vs. normal. Cell color is determined by the best gene rank percentile for the analyses within the cell. Red, up-regulated; blue, down-regulated. Numbers within each cell represented the number of analyses that met the threshold for the corresponding entry. Expression results in the liver cancer were highlighted in a green box.
Figure 2.
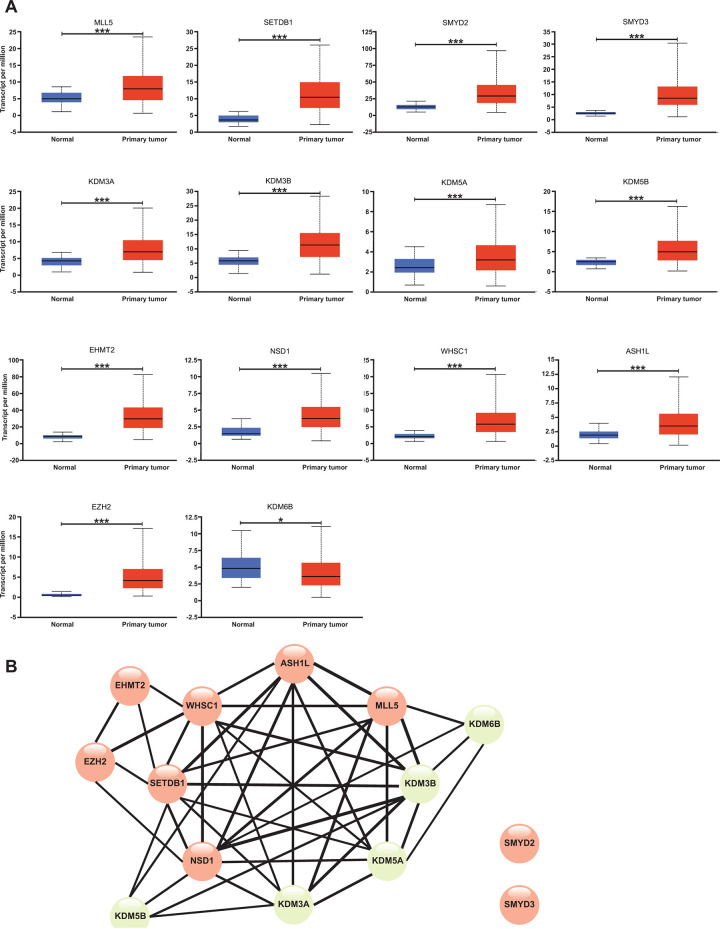
mRNA expression of 14 methyltransferases/demethylases between HCC and adjacent normal liver tissues, and co-occurrence analysis among these abnormal mRNA expression changes. (A) Relative mRNA expression difference was retrieved from UALCAN. n(Normal) = 50, and n(Primary tumor) = 371. (B) Co-occurrence results of mRNA expression changes in HCC were retrieved from ENCORI. Results with correlation coefficient (|r|) > 0.5 and p < 0.05 were included. Line thickness indicated the magnitude of the correlation coefficients (r). methyltransferases in red; demethylases in green. *p < 0.05, ***p < 0.001.
Co-Existence of the mRNA Expression Change in 14 Methyltransferases/Demethylases
From the above results, we found that 4 methyltransferases (MLL5, SMYD2, SMYD3, and ASH1 L) and 2 demethylases (KDM5A and KDM5B) for H3K4 were up-regulated in HCC (Figure 2A). However, the associations between these changes, especially within the same histone lysine, required further analysis. Therefore, we performed co-expression analyses to understand the associations through the ENCORI platform. Interestingly, we found that MLL5, ASH1 L, SETDB1, NSD1, WHSC1, KDM3A, KDM3B, and KDM5A were co-expressed with each other, indicating that the up-regulation of these genes might be induced through the same mechanism (Figure 2B). Meanwhile, the mRNA expression changes of SMYD2 and SMYD3 seemed to be independent of those of the remaining 12 genes. Even though multiple methyltransferases and demethylases for the same histone lysine were found to be up-regulated and partially co-expressed (for instance, SETDB1 co-expressed with KDM3A and KDM3B in H3K9), KDM6B, the only down-regulated demethylase in HCC (Figure 2A), did not correlate with EZH2 (r = 0.193, p-value = 1.70e-04), the only methyltransferase for H3K27, implying that an overall higher methylation level of H3K27 might be detected in HCC either by up-regulation of EZH2 or down-regulation of KDM6B. Together, the complicated associations reminded us of the importance of investigating multiple methyltransferases and demethylases for a specific histone lysine at the same time.
Correlation Between mRNA Expression Changes and Clinical Parameters of HCC Patients
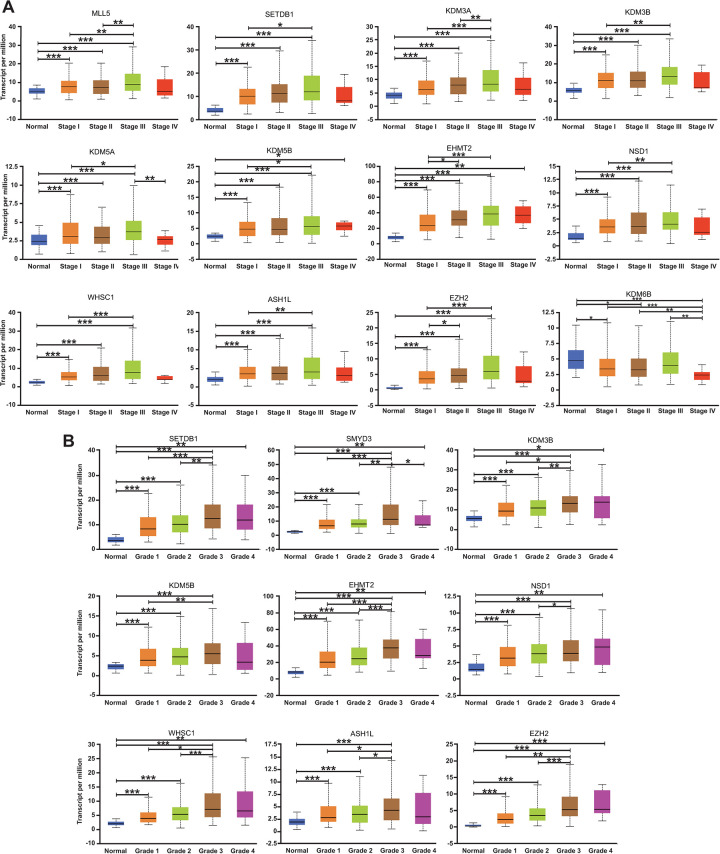
Clinical staging and tumor grading are important clinical parameters for HCC patients. We aimed to investigate the correlation between the aforementioned 14 methyltransferases/ demethylases with abnormal mRNA expression, and individual cancer stages, as well as tumor grades, in HCC patients by UALCAN. As was indicated in Figure 3A, the mRNA expression of 12 genes was significantly correlated with the individual cancer stages of HCC patients, among which 11 genes showed a positive correlation (the more advanced the clinical stage was, the higher the mRNA expression of specific genes tended to be) and only 1 gene, the down-regulated KDM6B, displayed a negative correlation. Meanwhile, the mRNA expression of these 11 genes peaked in stage 3 rather than in stage 4, at least partially owing to the limited sample size of patients in stage 4. The mRNA expression of KDM6B was noticeably the lowest in stage 4. Two remaining genes, SMYD2 and SMYD3, were found to not correlate with mRNA expression level and cancer stages (Fig. S2A). Similarly, the mRNA expression of 9 genes was found to be positively correlated with tumor grades of HCC patients, all peaking in grade 3 (Figure 3B). The remaining 5 genes showed no significant changes in mRNA expression levels within different grades of HCC (Fig. S2B). Together, we identified 12 genes closely correlated with cancer stages and 9 genes associated with tumor grades in HCC by UALCAN, among which 8 over-lapped genes (SETDB1, KDM3B, KDM5B, EHMT2, NSD1, WHSC1, ASH1 L, and EZH2) were obtained to imply a stronger association with the clinical parameter in HCC patients.
Figure 3.
mRNA expression of multiple methyltransferases/demethylases showing correlation with the clinical parameters of HCC patients (UALCAN). (A) Correlation between mRNA expression and the individual cancer stages. n(Normal) = 50, n(Stage I) = 168, n(Stage II) = 84, n(Stage III) = 82, and n(Stage IV) = 6. (B) Correlation between mRNA expression and the tumor grades. Grade 1, well differentiated (low grade); Grade 2, moderately differentiated (intermediate grade); Grade 3, poorly differentiated (high grade); Grade 4, undifferentiated (high grade). n(Normal) = 50, n(Grade 1) = 54, n(Grade 2) = 173, n(Grade 3) = 118, and n(Grade 4) = 12. Groups without statistical significance were unmarked. *p < 0.05, **p < 0.01, ***p < 0.001.
Four Methyltransferases/Demethylases Indicating Worse Overall Survival in HCC Patients
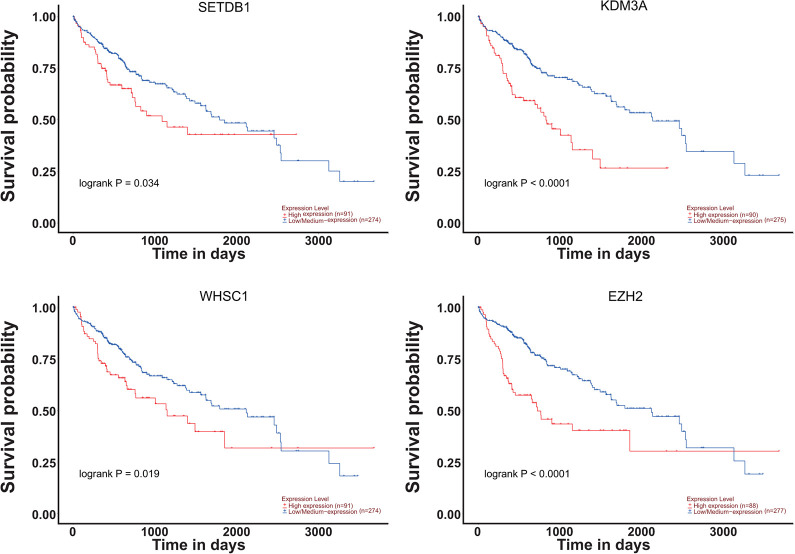
Additionally, we investigated the prognostic values of these 14 methyltransferases/ demethylases in HCC patients by plotting the correlation between the mRNA expression and the overall survival (OS) of patients. As was displayed in Figure 4, the elevated mRNA expression of SETDB1 (p = 0.034), KDM3A (p < 0.0001), WHSC1 (p = 0.019), EZH2 (p < 0.0001) was significantly correlated with shorter OS among patients with HCC, while mRNA expressions of the remaining 10 genes were found to be less indicative of prognostic values (Fig. S3). Noticeably, SETDB1, WHSC1, and EZH2 were correlated with clinical staging/grading of HCC in the above results as well, implying that they might be used as useful clinical biomarkers for the classification and prognosis prediction of HCC patients.
Figure 4.
Kaplan Meier plots showing significant effects of 4 methyltransferases/demethylases on overall survival of HCC patients (UALCAN). n = 365. High expression represented expression value > third quartile. p<0.05 was considered to be significant.
In short, by integrating the 8 over-lapped genes, that were associated with cancer stages and tumor grades, and 4 genes, that were correlated with the overall survival of HCC patients, we identified a total number of 9 methyltransferases/demethylases (SETDB1, KDM3A, KDM3B, KDM5B, EHMT2, NSD1, WHSC1, ASH1 L, and EZH2) that might play crucial roles in the development or the prognosis prediction of HCC via dysregulation of mRNA expression levels.
Genetic Alterations of 49 Methyltransferases/Demethylases in HCC
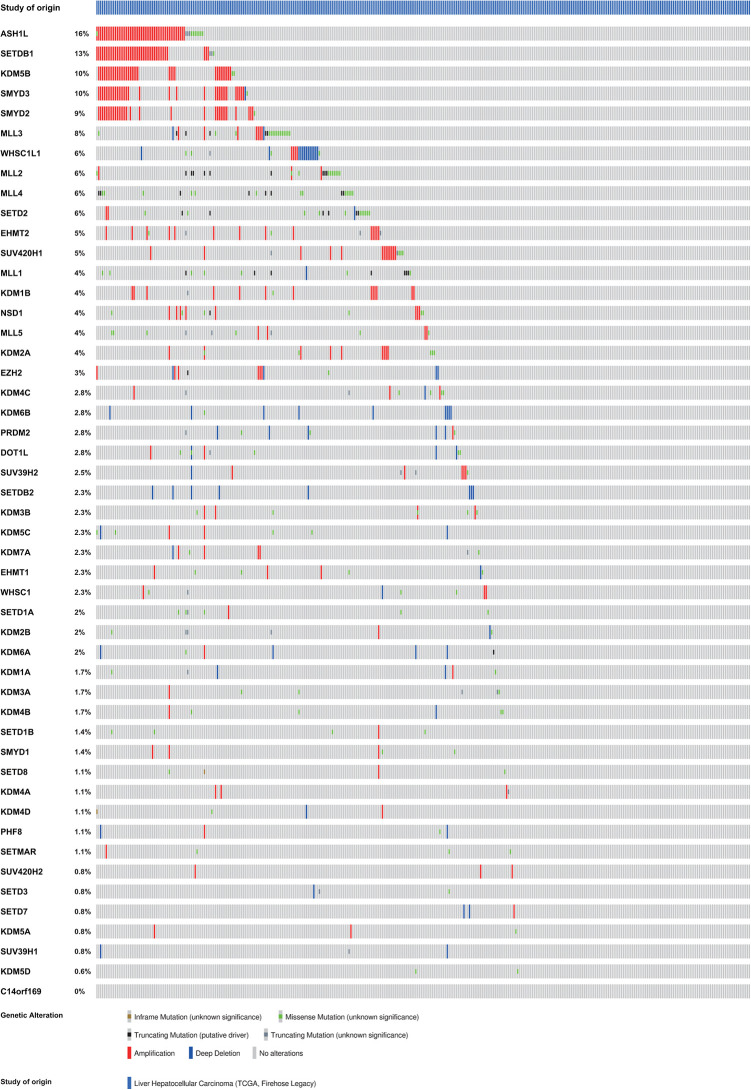
In addition to exploring the mRNA expression change of 49 methyltransferases/demethylases in HCC, we analyzed 2 major categories of genetic alterations in these genes, mutations, and copy number alterations (CNA). A group of 353 HCC patients was selected following our inclusion criteria in cBioPortal. ASH1 L was the most frequently altered one among the 49 genes, with an alteration rate of 16% among 353 samples, while RIOX1 was found to have no alteration at all (Figure 5). CNA (specifically, amplification) took a large proportion of the alterations in the top 5 genetically altered methyltransferases/demethylases (ASH1 L, SETDB1, KDM5B, SMYD3, and SMYD2), whereas missense mutations occurred more frequently in MLL3 (Figure 5). Following the threshold of alteration rate >5%, we collected the top 12 altered methyltransferases/demethylases for further studies.
Figure 5.
Mutations and copy number alterations of 49 methyltransferases/demethylases in patients with HCC (cBioPortal). n = 353.
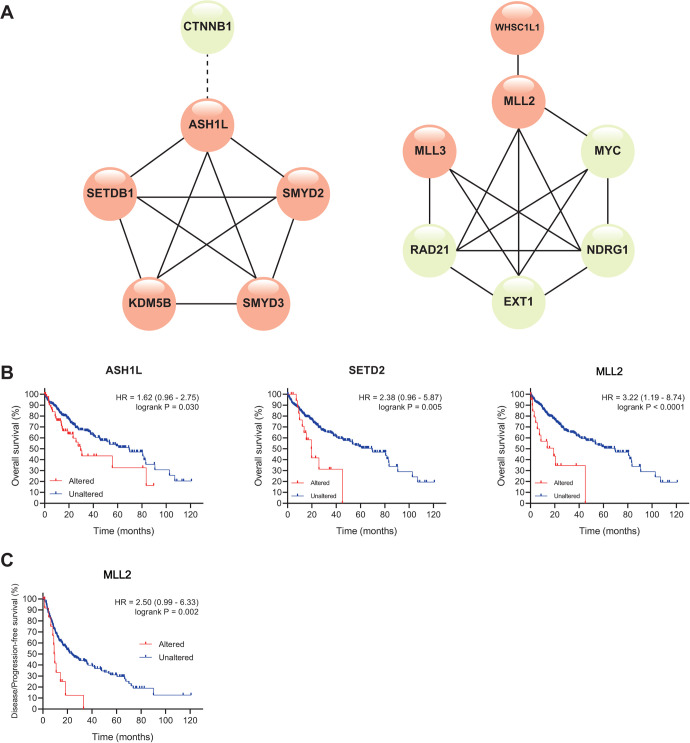
Co-occurrence Among the Top 12 Genetic Altered Methyltransferases/Demethylases and the Top 7 Altered Driver Genes in HCC
To investigate the relationship among these altered genes and their association with the top altered genes in HCC, we firstly identified 7 genetic altered candidate driver cancer genes in HCC, including mutations and CNAs. TP53, CTNNB1, and ALB were the only 3 genes that met our inclusion criteria for mutated genes in HCC patients, with a mutation frequency of 31.70%, 27.20%, and 11.60%, respectively (Table S2). Similarly, EXT1, RAD21, MYC, and NDRG1 were identified as the top 4 CNA candidate drive genes in HCC (Table S2). Next, we performed a mutual exclusivity/co-occurrence analysis among the top 12 altered methyltransferases/demethylases and the top 7 altered candidate driver cancer genes. Surprisingly, 5 genes with a larger alteration rate of amplification, ASH1 L, SETDB1, KDM5B, SMYD3, and SMYD2, were found to have co-occurrence with each other, indicating that patients bearing alterations of any one among these 5 genes were likely to possess the remaining 4 gene alterations (Figure 6A, left). However, these 5 genes had no co-occurrence with the alteration of the top 7 candidate driver cancer genes, and the alterations of ASH1 L were mutually exclusive from those of CTNNB1. Interestingly, the top 4 CNA candidate driver genes in HCC, EXT1, RAD21, MYC, and NDRG1, were co-occurred as well (Figure 6A, right). Genetic alterations in MLL3 co-existed with those in RAD21, EXT1, and NDRG1, while the co-occurrence of alterations in MLL2 was not only found in the above genes, but also in MYC and WHSC1L1. Detailed analysis information could be found in Table S3. These results provided a perspective for understanding the relationship among genetic alterations of methyltransferases/ demethylases and the possible mechanisms underlying such alterations.
Figure 6.
Co-occurrence analysis and prognostic analysis of top 12 altered methyltransferases/demethylases in HCC (cBioPortal). (A) Mutual exclusivity/co-occurrence analysis with top 7 altered candidate driver cancer genes. Solid lines, co-occurrence between 2 connected genes; dotted lines, mutual exclusivity between 2 connected genes. Genes in light red, altered methyltransferases/demethylases; Genes in light green, altered candidate driver cancer genes. Only significant results were shown (q-Value < 0.05). Correlation between genetic altered methyltransferases/demethylases and the overall survival (B) or disease/progression-free survival (C) of HCC patients.
Prognostic Values of 12 Genetic Altered Methyltransferases/Demethylases in HCC
Next, we evaluated the associations between genetic alterations of the top 12 altered methyltransferases/demethylases and OS and disease/progression-free survival (D/PFS) in HCC. Among 353 selected patient samples, patients with altered ASH1 L (HR = 1.62, p = 0.030), SETD2 (HR = 2.38, p = 0.005) and MLL2 (HR = 3.22, p < 0.0001) were associated with shorter OS of HCC patients (Figure 6B). Alterations in MLL2 showed the greatest and the most significant impact on OS, and it was the only alteration that led to shorter D/PFS in HCC patients (Figure 6C, HR = 2.50, p= 0.002). Non-significant associations could be found in Fig. S4. Together, we identified alterations in 3 genes, ASH1 L, SETD2, and MLL2, that might be of great prognostic value in HCC.
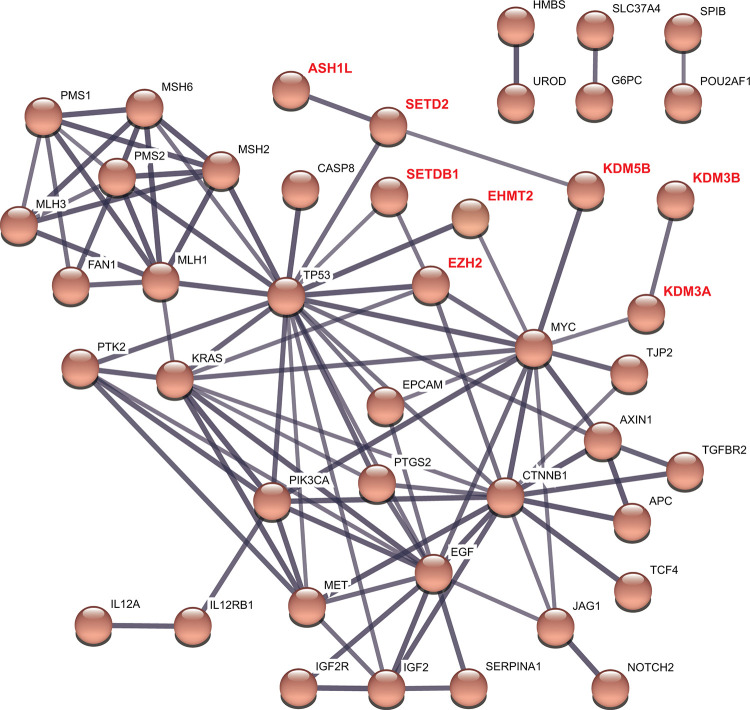
PPI Network of Important Methyltransferases/Demethylases and Seed Genes in HCC
From the above results, we obtained 9 methyltransferases/demethylases at the mRNA level (SETDB1, KDM3A, KDM3B, KDM5B, EHMT2, NSD1, WHSC1, ASH1 L, and EZH2) and 3 at the genetic level (ASH1 L, MLL2, SETD2) that were associated with clinical characteristics of HCC patients. A total number of 11 genes were therefore received as important methyltransferases and demethylases in HCC. Next, we associated these 11 genes in a PPI network with 50 seed genes identified in the Phenolyzer software (Table S4) to investigate their possible roles and mechanisms at the protein level. 44 nodes and 97 edges were displayed in the PPI network (Figure 7). 8 out of 11 important methyltransferases and demethylases were found to be associated with other genes in the network and distributed in 3 clusters. SETDB1 and SETD2 might be directly associated with TP53, while KDM3A and KDM5B might function through MYC. EHMT2 and EZH2 were both correlated with TP53 and MYC. Importantly, EZH2 also correlated with 2 other seed genes in the network, KRAS, and CTNNB1. The associations among methyltransferases/demethylases and seed genes might help understand the mechanisms and provide novel perspectives underlying the development of HCC.
Figure 7.
Protein-protein interaction of 11 important methyltransferases/demethylases and 50 seed genes in HCC (STRING). Line thickness indicated the strength of data support. The network was divided into 4 clusters by kmeans. Solid lines, correlation between 2 genes in the same cluster; dotted lines, correlation between 2 genes in different clusters.
Discussion
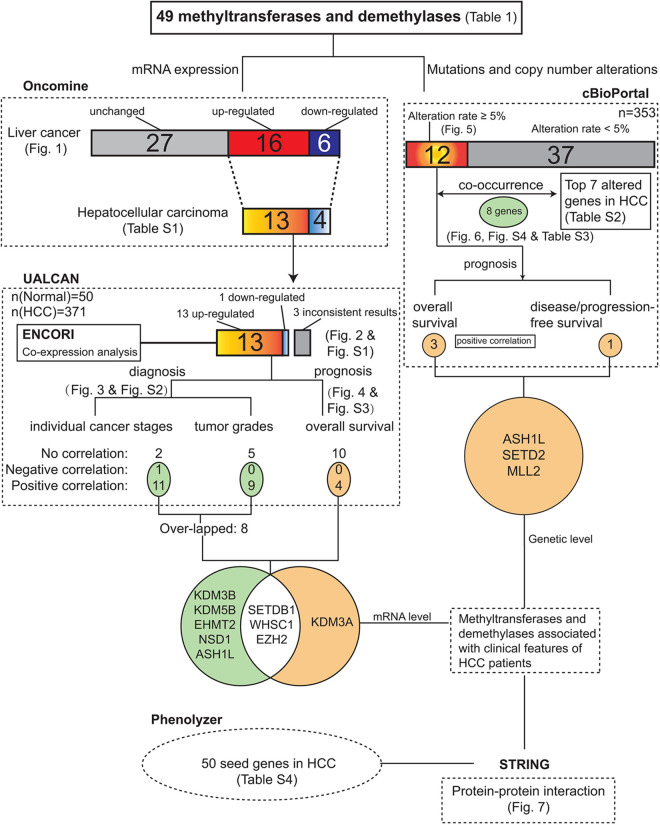
Accumulation of epigenetic alterations, including methylation of histone lysines, contributes to the development of HCC.3 Multiple methyltransferases and demethylases regulating the methylation of such histone lysines are indicated to play substantial roles in various cancers, including HCC.5,11,17 However, the associations among these enzymes and their distinct functions in HCC remain elusive. In this study, a comprehensive investigation of 49 methyltransferases and demethylases for H3K9, H3K9, H3K27, H3K36, H3K79, and H4K20 in HCC was performed with the help of varied integrative databases and platforms. A summary was shown in Figure 8.
Figure 8.
A summary regarding the associations and alterations of 49 methyltransferases and demethylases in HCC.
Firstly, we investigated the mRNA expression difference of 49 methyltransferases and demethylases between HCC tissues and normal liver tissues, and 13 up-regulated and 1 down-regulated genes were retrieved from the Oncomine and UALCAN databases (Figure 2A). Co-expression analysis of these 14 enzymes showed a complex co-expression network (Figure 2B), implying that changes in the expression of a certain enzyme might affect each other or that a common mechanism existed among the regulation of multiple enzymes. This also reminded us of the necessity to confirm the expression level of associated enzymes and the corresponding methylation status of the histone lysine, since these enzymes might work co-operatively at the same methylation site. However, it was also noted that not all associated enzymes would lead to the same pathological or clinical outcome in HCC. Clinical staging and pathological grading were 2 crucial clinical indicators of HCC. Correlation analysis revealed that 8 out of 14 enzymes were correlated with clinical stages and pathological grades (Figure 3). Furthermore, the higher mRNA expression of SETDB1, KDM3A, WHSC1, and EZH2 was found to be associated with shorter OS of HCC patients (Figure 4). Intriguingly, the expression of these enzymes was all associated with that of SEDB1 and WHSC1 (Figure 2B), making both genes promising research hotpots in the development and prediction of prognosis in HCC.
SETDB1, an enzyme that methylates H3K9, is considered to play important roles in multiple cancer types, including acute myeloid leukemia, melanoma, and HCC.18-21 Guo et al. reported that methylation by SETDB1 activated AKT and could act as an oncogenic marker.22 In HCC, over-expression of SETDB1, induced by multiple complementary mechanisms, was correlated with proliferation and progression.23,24 And it was suggested that SETDB1 played a crucial role in the metastatic progression of HCC.24 Additionally, Shao et al. found that the p53-signaling pathway could be activated by miR-621 via inhibition of SETDB1 to improve the radiosensitivity of HCC cells.21 On the other hand, H3K36me2 has been regarded as the principal product of WHSC1 and this regulation of methylation is sufficient for gene activation. Over-expression of WHSC1 has been reported in multiple cancer types, including multiple myeloma,25 bladder cancer, and lung cancer,26 Alterations of H3K36me2 caused by WHSC1 were demonstrated to promote the initiation of oncogenic programming through activation of several pathways, such as the Wnt pathway.26,27 Zhang et al. reported that WHSC1-mediated dimethylation of PTEN regulated the repair of DNA double-strand breaks, suggesting a promising therapeutic approach by inhibiting WHSC1 in cancer treatment.28 In prostate cancer, WHSC1 was identified as a driver of metastatic progression, and silencing of WHSC1 by MCTP-39 led to reduced expression of H3K36me2 and decreased tumor volume in prostate cancer xenografts.29 Moreover, another report found that WHSC1 linked a vicious signaling cascade between AKT and RICTOR to facilitate unrestrained AKT signaling in a PTEN-null murine prostate cancer model, driving tumor metastasis.30 Feng et al. addressed that WHSC1 was recruited to the estrogen receptor alpha (ERα) gene to promote the expression of ERα in breast cancer, providing a novel regulating mechanism that might become a promising therapeutic target for patients with resistance to tamoxifen.31 Even though the detailed effect and mechanism of WHSC1 in HCC remain elusive, over-expression of WHSC1 might participate in the tumorigenesis of non-alcoholic steatohepatitis related hepatocarcinogenesis.32
Secondly, we determined the mutation and copy number alteration status of 49 methyltransferases and demethylases in HCC, and 12 enzymes showing a genetic alteration rate higher than 5% were retrieved (Figure 5). Interestingly, amplification was the major genetic alteration found in the top 5 altered enzymes (ASH1 L, SETDB1, KDM5B, SMYD3, and SMYD2) in HCC, and these alterations were co-existed with each other (Figure 6A), implying that a shared mechanism might control such changes. It was also reported that 4 HMTs (SETDB1, SMYD3, ASH1 L, SMYD2) with genetic alterations were regarded as candidate therapeutic targets in breast cancer,33 and the reason KDM5B was not involved might be the fact that it was a demethylase and not covered in this study. This reminded us that the co-existence of these 5 genetic alterations might be an important process in the initiation and development of cancer. Are they functionally dependent or key points under the same mechanism? What impact does their co-expression have on HCC? Is the alteration of KDM5B, as the only demethylase among these 5, a compensatory change or a temporal change at different stages of tumorigenesis and development? Does it represent a particular type of HCC? These questions require further investigation. Combined with the results that RNA expression of ASH1 L, SETDB1, and KDM5B was positively correlated, while that of SMYD2 and SMYD3 had little association (Figure 2B), it might be speculated that amplification of ASH1 L, SETDB1, and KDM5B at least partially contributed to the elevation of their mRNA expressions and that there might be a different mechanism regulating the transcription of SMYD2 and SMYD3. MLL2 mutation is a driver in various cancer types, but the mechanism remains elusive. Missense alterations in MLL2 and MLL3 were co-expressed with CNA alterations in 4 HCC driver genes (MYC, RAD21, EXT1, and NDRG1) (Figure 6A), suggesting the importance of MLL2 and MLL3 in HCC development. Ding et al. reported that MLL2 was a direct target of TP53R249S 34, while it was suggested that MLL mutation resulted in genome instability, which could help explain its widespread role in tumorigenesis.35 Even though the frequency of MLL2 mutations is not high in HCC, HCC patients with these alterations have a shorter OS (HR = 3.22) and shorter DFS/PFS (HR = 2.50), suggesting a poor clinical prognosis (Figure 6B&C). Therefore, the detection of MLL2 mutations and the study of the related mechanism of the role of MLL2 mutation in the initiation and development of HCC should be warranted.
An important study highlighting the complex molecular network of H3K4 KMT enzymes and tri-demethylases that regulate site-specific copy gains can be referred elsewhere.36 Even though we are still taking baby steps in exploring this complex association network of genetic alterations among methyltransferases and demethylases, some important findings for each enzyme have been indicated. A whole-genome mutational analysis by Fujimoto et al. recognized ASH1 L and SETDB1 as novel driver genes for liver cancer.37 ASH1 L mediates the dimethylation of H3K36 and trimethylation of H3K4.38 While methylation of H3K4 mediated by ASH1 L was found to be crucial in the global-genome nucleotide excision repair activity to prevent ultraviolet light-induced skin cancer,39 the ASH1L-mediated dimethylation mark of H3K36 could be recognized by LEDGF to recruit mixed-lineage leukemia (MLL)-associated proteins to activate gene expression of principle targets in leukemia cells, the process of which could be specifically antagonized by KDM2A, the demethylation of H3K36.40 Therefore, the interactions of ASH1L-LEDGF-KDM2A were considered crucial underlying the tumorigenesis of MLL-associated leukemia.40 Meanwhile, KDM5B, a demethylase for H3K4, was also found to negatively regulate the leukemogenesis of MLL-rearranged AML cells, proposing a key role for the (de)methylation of H3K4 in determining leukemia stem cell fate.41 KDM5B participates in the inhibition of stimulator of interferon genes (STING) through demethylation of H3K4, and thus plays a role in the natural immune defense against microbial infection and cancer.42 In ER+ luminal breast cancer, KDM5B regulates the heterogeneity of cellular transcriptome, rather than the genetic heterogeneity, and its higher expression due to amplification and/or overexpression is correlated with worse OS.43,44 While the roles of ASH1 L in HCC remain elusive, the up-regulation of KDM5B has been frequently observed in HCC, and depletion of KDM5B suppressed the proliferation and invasion of HCC cells partly through up-regulation of p15 and p27, making KDM5B a potential therapeutic target in HCC patients.45,46 Both MLL2 (originally referred to as MLL4) and MLL3 are members of the COMPASS family of H3K4 methyltransferases. MLL2-mediated H3K4me3 has been reported to be crucial for a specific subset of genes through binding to non-TSS regulatory elements during embryonic development.47 In cancer, there have been 2 models proposed for the implications of mutations in MLL2/MLL3. The first model proposed by Herz et al. indicated that somatic mutations in MLL2/MLL3 could result in gain-of-function on the enhancers of oncogenes or loss-of-function on the enhancers of tumor suppressor genes, thus contributing to the development of cancer.48 The second model proposed that MLL2 mutations could lead to genomic instability and increase gene mutations, facilitating the heterogeneity, and promoting tumorigenesis.35 Therefore, the associations between mutations in MLL2/MLL3 and top altered candidate driver cancer genes in HCC might be explained by such models. However, further investigation regarding this matter in HCC was needed.
A total number of 11 methyltransferases/demethylases were identified to be associated with the clinical characteristics of HCC patients, 9 from the mRNA expression level and 3 from the genetic alteration level (ASH1 L was identified in both levels). These enzymes might serve as potential prognostic biomarkers for HCC. The indicated associations between these methyltransferases/demethylases and seed genes of HCC in the PPI network raised numerous potential research questions in the field of HCC (Figure 7). For example, what is the association between SETDB1 and EZH2 in HCC, considering that both enzymes have been proved to be important? Is it concomitant or cause-and-effect? Additionally, the phenomenon that EZH2 correlated with 4 known driver genes in HCC (TP53, KRAS, MYC, and CTNNB1) reminded us of the important roles EZH2 might play in the development of HCC. Given the complex network within methyltransferases and demethylases, research into the combined effects of associated enzymes in HCC should be guaranteed.
There are some limitations to this study. The sample sources, sample background information, and algorithms of various databases are inconsistent at present; lack of a unified and standardized process may lead to inconsistent results when analyzing across databases or studies; dependence on existing data and results may lead to some “false negative” results; lack of further experimental verification. Despite these limitations, we have identified methyltransferases/demethylases that may play an important role in HCC through the joint analysis of multiple databases in this study, and we believe that this study can provide potential targets in the future investigation of HCC.In general, we analyzed 49 major methyltransferases/demethylases for H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20 in HCC from the mRNA level and the genetic level using multiple datasets and databases. 11 of them were associated with clinical characteristics of HCC patients and might serve as promising prognostic markers in HCC. This study provided a general understanding of the associations between these methyltransferases/demethylases in HCC. Therefore, further investigation of the precise roles and mechanisms underlying these associations in HCC is promising.
Supplemental Material
Supplemental Material, Supplementary_Figure_legends for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Fig._S1 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Fig._S2 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Fig._S3 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Fig._S4 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Table_S1 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Table_S2 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Table_S3 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Table_S4 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Abbreviations
- CNAs
copy number alterations
- D/PFS
disease/progression-free survival
- Erα
estrogen receptor alpha
- HCC
hepatocellular carcinoma
- H3K4
histone H3 at lysine 4
- H3K9
histone H3 at lysine 9
- H3K27
histone H3 at lysine 27
- H3K36
histone H3 at lysine 36
- H3K79
histone H3 at lysine 79
- H4K20
histone H4 at lysine 20
- KDMs
histone lysine demethylases
- KMT
histone lysine methyltransferase
- OS
overall survival
- STING
stimulator of interferon genes
Footnotes
Ethical Statement: Our study did not require an ethical board approval because it did not contain human or animal trials.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the Jilin Province Department of Finance (2018SCZWSZX-020) and the Jilin Province Department of Science and Technology (20170623009TC).
Supplemental Material: Supplementary material for this article is available online.
ORCID iD: Ziling Liu  https://orcid.org/0000-0003-0582-7579
https://orcid.org/0000-0003-0582-7579
References
- 1. Henley SJ, Ward EM, Scott S, et al. Annual report to the nation on the status of cancer, part I: national cancer statistics. Cancer. 2020;126(10):2225–2249. doi:10.1002/cncr.32802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi:10.1016/s0140-6736(18)30010-2 [DOI] [PubMed] [Google Scholar]
- 3. Villanueva A. Hepatocellular carcinoma. New Engl J Med. 2019;380(15):1450–1462. doi:10.1056/NEJMra1713263 [DOI] [PubMed] [Google Scholar]
- 4. Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62(6):394–399. doi:10.3322/caac.21161 [DOI] [PubMed] [Google Scholar]
- 5. Au SL, Ng IO, Wong CM. Epigenetic dysregulation in hepatocellular carcinoma: focus on polycomb group proteins. Front Med. 2013;7(2):231–241. doi:10.1007/s11684-013-0253-7 [DOI] [PubMed] [Google Scholar]
- 6. Hasegawa K, Kokudo N, Makuuchi M, et al. Comparison of resection and ablation for hepatocellular carcinoma: a cohort study based on a Japanese nationwide survey. J Hepatol. 2013;58(4):724–729. doi:10.1016/j.jhep.2012.11.009 [DOI] [PubMed] [Google Scholar]
- 7. Ishizawa T, Hasegawa K, Aoki T, et al. Neither multiple tumors nor portal hypertension are surgical contraindications for hepatocellular carcinoma. Gastroenterology. 2008;134(7):1908–1916. doi:10.1053/j.gastro.2008.02.091 [DOI] [PubMed] [Google Scholar]
- 8. Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13(2):123–135. doi:10.1038/nrc3449 [DOI] [PubMed] [Google Scholar]
- 9. Cornett EM, Ferry L, Defossez PA, Rothbart SB. Lysine methylation regulators moonlighting outside the epigenome. Mol Cell. 2019;75(6):1092–1101. doi:10.1016/j.molcel.2019.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Husmann D, Gozani O. Histone lysine methyltransferases in biology and disease. Nat Struct Mol Biol. 2019;26(10):880–889. doi:10.1038/s41594-019-0298-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Højfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Disc. 2013;12(12):917–930. doi:10.1038/nrd4154 [DOI] [PubMed] [Google Scholar]
- 12. Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. doi:10.1016/j.molcel.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rea S, Eisenhaber F, O’Carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–599. doi:10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- 14. The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi:10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu H, Liu Y, Liu W, Zhang W, Xu J. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat Commun. 2015;6:8494 doi:10.1038/ncomms9494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fei Q, Shang K, Zhang J, et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat Commun. 2015;6:8651 doi:10.1038/ncomms9651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer. 2015;15(6):334–346. doi:10.1038/nrc3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robbez-Masson L, Tie CHC, Rowe HM. Cancer cells, on your histone marks, get SETDB1, silence retrotransposons, and go! J Cell Biol. 2017;216(11):3429–3431. doi:10.1083/jcb.201710068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cuellar TL, Herzner AM, Zhang X, et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemia. J Cell Biol. 2017;216(11):3535–3549. doi:10.1083/jcb.201612160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ceol CJ, Houvras Y, Jane-Valbuena J, et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471(7339):513–517. doi:10.1038/nature09806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shao Y, Song X, Jiang W, et al. MicroRNA-621 acts as a tumor radiosensitizer by directly targeting SETDB1 in hepatocellular carcinoma. Mol Ther. 2019;27(2):355–364. doi:10.1016/j.ymthe.2018.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo J, Dai X, Laurent B, et al. AKT methylation by SETDB1 promotes AKT kinase activity and oncogenic functions. Nat Cell Biol. 2019;21(2):226–237. doi:10.1038/s41556-018-0261-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y, Huang J, Li Q, et al. Histone methyltransferase SETDB1 promotes cells proliferation and migration by interacting withTiam1 in hepatocellular carcinoma. BMC Cancer. 2018;18(1):539 doi:10.1186/s12885-018-4464-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wong CM, Wei L, Law CT, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63(2):474–487. doi:10.1002/hep.28304 [DOI] [PubMed] [Google Scholar]
- 25. Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92(9):3025–3034. [PubMed] [Google Scholar]
- 26. Toyokawa G, Cho HS, Masuda K, et al. Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia. 2011;13(10):887–898. doi:10.1593/neo.11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuo AJ, Cheung P, Chen K, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell. 2011;44(4):609–620. doi:10.1016/j.molcel.2011.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang J, Lee YR, Dang F, et al. PTEN methylation by NSD2 controls cellular sensitivity to DNA damage. Cancer Disc. 2019;9(9):1306–1323. doi:10.1158/2159-8290.Cd-18-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aytes A, Giacobbe A, Mitrofanova A, et al. NSD2 is a conserved driver of metastatic prostate cancer progression. Nat Commun. 2018;9(1):5201 doi:10.1038/s41467-018-07511-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li N, Xue W, Yuan H, et al. AKT-mediated stabilization of histone methyltransferase WHSC1 promotes prostate cancer metastasis. J Clin Investig. 2017;127(4):1284–1302. doi:10.1172/jci91144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng Q, Zhang Z, Shea MJ, et al. An epigenomic approach to therapy for tamoxifen-resistant breast cancer. Cell Res. 2014;24(7):809–819. doi:10.1038/cr.2014.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuramoto J, Arai E, Tian Y, et al. Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis. Carcinogenesis. 2017;38(3):261–270. doi:10.1093/carcin/bgx005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu L, Kimball S, Liu H, Holowatyj A, Yang ZQ. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer. Oncotarget. 2015;6(4):2466–2482. doi:10.18632/oncotarget.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ding X, He M, Chan AWH, et al. Genomic and epigenomic features of primary and recurrent hepatocellular carcinomas. Gastroenterology. 2019;157(6):1630–1645.e6. doi:10.1053/j.gastro.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 35. Kantidakis T, Saponaro M, Mitter R, et al. Mutation of cancer driver MLL2 results in transcription stress and genome instability. Genes Dev. 2016;30(4):408–420. doi:10.1101/gad.275453.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mishra S, Van Rechem C, Pal S, et al. Cross-talk between lysine-modifying enzymes controls site-specific DNA amplifications. Cell. 2018;174(4):803–817.e16. doi:10.1016/j.cell.2018.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–509. doi:10.1038/ng.3547 [DOI] [PubMed] [Google Scholar]
- 38. Vizoso M, Esteller M. The activatory long non-coding RNA DBE-T reveals the epigenetic etiology of facioscapulohumeral muscular dystrophy. Cell Res. 2012;22(10):1413–1415. doi:10.1038/cr.2012.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Balbo Pogliano C, Gatti M, Rüthemann P, Garajovà Z, Penengo L, Naegeli H. ASH1 L histone methyltransferase regulates the handoff between damage recognition factors in global-genome nucleotide excision repair. Nat Commun. 2017;8(1):1333 doi:10.1038/s41467-017-01080-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu L, Li Q, Wong SH, et al. ASH1 L links histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Disc. 2016;6(7):770–783. doi:10.1158/2159-8290.Cd-16-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wong SH, Goode DL, Iwasaki M, et al. The H3K4-methyl epigenome regulates leukemia stem cell oncogenic potential. Cancer Cell. 2015;28(2):198–209. doi:10.1016/j.ccell.2015.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu L, Cao J, Cai WL, et al. KDM5 histone demethylases repress immune response via suppression of STING. PLoS Biol. 2018;16(8):e2006134 doi:10.1371/journal.pbio.2006134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hinohara K, Wu HJ, Vigneau S, et al. KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance. Cancer Cell. 2018;34(6):939–953.e9. doi:10.1016/j.ccell.2018.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamamoto S, Wu Z, Russnes HG, et al. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell. 2014;25(6):762–777. doi:10.1016/j.ccr.2014.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang X, Oishi N, Shimakami T, et al. Hepatitis B virus X protein induces hepatic stem cell-like features in hepatocellular carcinoma by activating KDM5B. World J Gastroenterol. 2017;23(18):3252–3261. doi:10.3748/wjg.v23.i18.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang D, Han S, Peng R, et al. Depletion of histone demethylase KDM5B inhibits cell proliferation of hepatocellular carcinoma by regulation of cell cycle checkpoint proteins p15 and p27. J Exp Clin Cancer Res. 2016;35:37 doi:10.1186/s13046-016-0311-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu D, Gao X, Cao K, et al. Not all H3K4 methylations are created equal: Mll2/COMPASS dependency in primordial germ cell specification. Mol Cell. 2017;65(3):460–475. e6 doi:10.1016/j.molcel.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Herz HM, Hu D, Shilatifard A. Enhancer malfunction in cancer. Mol Cell. 2014;53(6):859–866. doi:10.1016/j.molcel.2014.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Supplementary_Figure_legends for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Fig._S1 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Fig._S2 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Fig._S3 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Fig._S4 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Table_S1 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Table_S2 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Table_S3 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment
Supplemental Material, ZHENG_Table_S4 for Comprehensive Bioinformatics Analysis of Key Methyltransferases and Demethylases for Histone Lysines in Hepatocellular Carcinoma by Yang Zheng, Lili Tang, Guojiang Chen and Ziling Liu in Technology in Cancer Research & Treatment