Abstract
The cerebral cortex contains a significant quantity of intracortical myelin, but the genetics of cortical myelination (CM) in humans is not well understood. Relatively novel MRI-derived measures now enable the investigation of cortical myelination in large samples. In this study, we use a genetically-informative neuroimaging sample of 1,096 young adult subjects from the Human Connectome Project in order to investigate genetic and environmental variation in CM and its relationships with cerebral surface area (SA) and cortical thickness (CT). We found that genetic factors account for approximately 50% of the observed individual differences in mean cortical myelin, 75% of the variation in total SA, and 85% of the variance in global mean CT. Although significant genetic influences were found throughout the cortex, both CM and SA demonstrated a posterior predominance, with disproportionately strong effects in the parietal and occipital lobes and significantly overlapping heritability maps (p<0.001). Yet despite showing similar spatial heritability patterns, we found evidence that CM is genetically independent from SA at both global and vertex levels; genetically-mediated relationships between CM and CT were similarly small in magnitude. We also found small but statistically significant genetic associations between NIH Toolbox Total Cognition score and CM in the temporal lobe and insula. SA-cognition and CT-cognition correlations were less widespread compared to CM and both patterns were similar to those reported in prior studies.
Keywords: Cortical myelination, surface area, cortical thickness, intelligence, genetics, MRI
Introduction
The human cerebrum is an extraordinarily complex structure whose successful construction involves a broad array of neurodevelopmental processes including neurogenesis, synaptogenesis, axonal pruning, and myelination, all of which are affected by a myriad of genetic and nongenetic influences. The advent of magnetic resonance imaging (MRI) has enabled the large-scale investigation of typical human brain structure in vivo. To date, imaging genetic studies have investigated numerous structural phenotypes at high resolution and have found that most are both heritable and topographically variable (Eyler et al., 2012; McKay et al., 2014; Rimol et al., 2010). The heritability of cortical thickness (CT) increases in childhood and adolescence (Schmitt et al., 2014), and younger adults is highest in the posterior frontal, parietal, and superior temporal lobes (Joshi et al., 2011; McKay et al., 2014). Twin and family studies on cerebral surface area (SA) in children and young adults have found that the posterior cerebrum, particularly the parasagittal occipital lobe and precuneus, are reproducibly the most influenced by genetic factors (Jha et al., 2018; Schmitt et al., 2019a; Strike et al., 2018; Yoon et al., 2012).
Cortical myelination (CM) represents a relatively novel noninvasive phenotype that is also derived from structural MRI data. The posterior cerebrum tends to be highly myelinated in humans, with similar posterior-predominant patterns to regions with the highest surface area (Glasser and Van Essen, 2011). Although the majority of white matter fibers lie below the cortical surface, many cortical regions are heavily myelinated. Using the inherent contrast within T1 and T2-weighted images, cortical myelin content can be estimated. These myelination maps have a high concordance with known myeloarchitectural and cytoarchitectural divisions (Glasser and Van Essen, 2011). Although myelination has been shown to be associated with cognitive ability and has a hypothesized role in schizophrenia (Grydeland et al., 2013; Whitford et al., 2012), the genetics of CM is not well-understood. The only prior imaging genetic study on CM found a strong anterior-posterior gradient in genetic influences strongest near the occipital pole (Liu et al., 2018), with overall pattern similar to heritability maps of SA. Surface area and intracortical myelination have been hypothesized to be neurodevelopmentally and evolutionarily coupled (Cafiero et al., 2019; Hill et al., 2010), raising the possibility of common genetic influences on these two phenotypes. Myelination has also been associated with cortical thinning of association areas in adolescence, and these two metrics may also be neurodevelopmentally coupled (Vandekar et al., 2015; Whitaker et al., 2016).
In the current study, we perform a systematic examination of the genetics of cortical myelination, cerebral surface area, and thickness at high spatial resolution in a younger adult sample. In addition to examining these measures separately, we also investigate them together via genetically-informative bivariate models and identify genetically-mediated correlates with cognition.
Methods
Data were obtained from the Human Connectome Project (HCP) S1200 release (Van Essen et al., 2012). This dataset includes high-resolution MRI structural neuroimaging data on 1,113 adults as well as a broad cognitive battery. The image acquisition protocol included T1 weighted MP-RAGE (TR 2400ms, TE 2.14ms, flip angle=8°, FOV 224 × 224 mm2, voxel size= 0.7 mm isotropic, scan time =7:40 min) and T2 weighted T2-SPACE (TR=3200 ms, TE=565ms, FOV 224 × 224 mm2, voxel size= 0.7 mm isotropic, scan time=8:24 min). All data were acquired on the same 3T scanner. Raw images were post-processed with using standard HCP pipelines; these pipelines have been described in detail elsewhere (Glasser et al., 2013). Briefly, the initial pipeline (‘PreFreesurfer’) aims to correct image distortion, isolate brain parenchyma from other tissues, and register images to a shared space. Vertex-level measures of cerebral surface area and cortical thickness were then calculated via FreeSurfer 5.3.0 (Fischl, 2012).
Cortical myelin content was estimated by calculating the ratio of T1-weighted to T2-weighted images; this ratio accentuates the inherent myelin contrast in both sequences while simultaneously attenuating the effects of magnetic field inhomogeneity (Glasser and Van Essen, 2011). It is noteworthy that T1/T2 ratios are not a direct measure of myelination but rather a convenient proxy; HCP provides unitless vertex-level estimates of myelination percentages. Estimates of cortical myelination using T1/T2 ratios are advantageous given their high spatial resolution, relatively short acquisition times, and high test-retest reliability (Arshad et al., 2017). Limitations include scaling discrepancies between MRI scanners (Ganzetti et al., 2014), potential dependence on iron or water content (Cafiero et al., 2019), and a generally lower validity compared to gold standard methods such as myelin water fraction (MWF) (Arshad et al., 2017; Uddin et al., 2018). Nevertheless, T1/T2 myelin mapping is still considered a useful (albeit imperfect) method for estimating myelin content (Shafee et al., 2015).
Post processing of brain measures involved image downsampling (we used HCP’s 32k vertex Conte 69 data), multimodal surface matching (MSM) which incorporates both structural and functional data for registration (Robinson et al., 2014), and conversion to the GIFTI file format. Image acquisition and image processing was performed using standard HCP guidelines (Marcus et al., 2013). Each subject’s myelination, surface area, and thickness maps were then smoothed with a 5 mm kernel via the ‘cifti-smoothing’ command from the Connectome Workbench (Van Essen et al., 2012).
Vertex-level measures were available for a total of 1,096 subjects (500 males, 596 females). The final sample included 229 subjects ages 22–25 years, 478 from 26–30, 376 ages 31–35, and 13 subjects over 35 years. Data from 442 families with up to 6 individuals per family were available, including families with twin-pairs. Genotype-derived zygosity information were available for 283 monozygotic (MZ) and 168 dizygotic (DZ) twins. An additional 66 self-reported MZ and 65 DZ twins also were included; we preferentially used genomic data to assign zygosity if available, but also included subjects with self-reported zygosity in order to maximize power. 349 MZ twins, 233 DZ twins, and 514 singletons were included in total. Intelligence was assessed via the NIH Toolbox’s ‘Total Cognition’ composite score (NTC); this score correlates highly (r=0.95) with traditional constructs of cognition in adults (Heaton et al., 2014) and is heritable in the HCP dataset (Schmitt et al., 2019b). The study was approved by the Institutional Review Board of the Hospital of the University of Pennsylvania.
Statistical Analysis
Each subject’s scalar neuroanatomic measures were imported into the R statistical environment for analysis (R Core Team, 2018) using the ‘cifti’ package and subsequently merged with demographic and cognitive data. The data were reformatted such that each record represented family-wise (rather than individual-wise) data. Genetic modeling was performed in OpenMx, a structural equation modeling package fully integrated into the R environment (Boker et al., 2011; Neale et al., 2016). First, univariate analyses of global CM (i.e. mean over all vertices), global mean CT, and total SA were performed via the classic ACE model with an extended twin design (Posthuma and Boomsma, 2000). This model decomposes the observed phenotypic variance into components attributable to additive genetic (A), shared environmental (C), and unique environmental factors (E) including measurement error (Lenroot et al., 2009; Neale and Cardon, 1992). Mathematically, these variance components can be estimated based on the observed phenotypic variance and cross-twin or cross-sibling covariances. For example:
Where VP represents the observed phenotypic variance, CovMZ the monozygotic twin-twin phenotypic covariance and CovDZ the dizygotic phenotypic covariance. From these three linear equations, the variance attributable to additive genetic factors (A) can be estimated, as well as estimates for the shared (C) and unique (E) environmental variance. Proportional variance estimates (e.g. the heritability, A/VP, or a2) can subsequently be calculated. We then used similar models to examine CM, SA, and CT on the vertex level. The classic extended twin design assumes equal shared environment between twins and non-twin siblings. In order to relax this assumption, we reanalyzed the data using ACET models, which additionally allow for variance owed to twin-specific environmental factors. As an alternative means to address this issue, we ran classic ACE models on a twins-only subset of the S1200 HCP data (N=582). The results of both ACET and twins-only analyses were similar to our original analyses and are presented in supplementary figures S1 and S2.
All models contained parameters to control for the effects of sex and age on the mean. Model fit was determined using maximum likelihood (Edwards, 1972). In order to test for statistical significance of individual variance components, fit of the full ACE model was compared to submodels with either genetic or shared environmental parameters removed (CE and AE models, respectively); differences in model fit asymptotically follow a 50:50 mixture of zero and χ2 with 1 degree of freedom (Dominicus et al., 2006). Control for multiple testing was performed with the false discovery rate (Genovese et al., 2002).
We then examined SA and CM simultaneously. In order to facilitate visual comparisons of heritability maps, we first constructed simple concordance maps by calculating the 10th, 25th, 50th, 75th, and 90th centiles for SA and CM heritability separately and then identified vertices where both metrics were above or below a given threshold (e.g. vertices with a2 above the 90th centile for both SA and CM). To evaluate spatial patterns more quantitatively, we then compared SA and CM heritability maps via spatial permutation, also referred to as the “spin” test (Alexander-Bloch et al., 2018; Vandekar et al., 2015). Briefly, cross-vertex Pearson’s correlations between a2 maps were plotted against a null distribution that was described by 1000 spatially-permuted values. This test is advantageous as it controls for both multiple testing and spatial autocorrelations. These analyses were repeated for pairwise comparisons with CT.
Shared genetic and nongenetic factors between SA and CM were then directly tested using genetically-informative bivariate Cholesky decomposition. Given the negligible role of the shared environment in the univariate models, it was not included in these bivariate models. Cholesky decomposition factors any symmetric positive definite matrix into a lower triangular matrix postmultiplied by its transpose (Neale and Cardon, 1992). Mathematically, the 2 × 2 phenotypic variance-covariance matrix (P), and expected cross-twin variance-covariance matrices (CovMZ, CovDZ) can be expressed as:
Where A and E represent 2 × 2 lower triangular matrices with 3 free parameters each, e.g.:
Similar to the univariate case, the observed cross-sibling variance-covariance matrices can be used to solve for each individual parameter estimate. Genetic and environmental correlations between SA and CM were calculated:
Where represents the genetic covariance between SA and CM at the ith, vertex and and the vertex-level genetic variance in CM and SA, respectively. Similar bivariate models were constructed for pairwise comparisons with CT.
Finally, we investigated brain-intelligence relationships by using similar bivariate models to quantify the covariance between vertex-level CM and NTC. In these models, NTC remained fixed to a scalar value for each subject, but CM was iterated at every vertex over the cerebrum. SA-NTC and CT-NTC covariances were assessed similarly.
Results
Univariate Heritability Maps
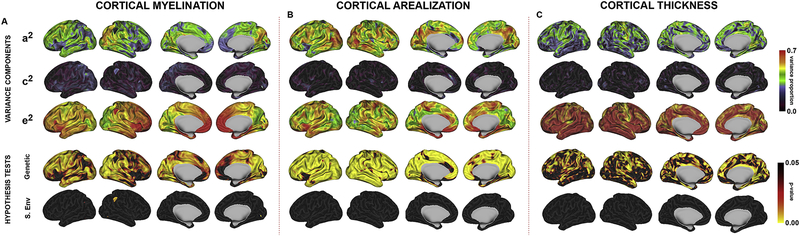
Global CM was a moderately heritable trait, with approximately half of the total phenotypic variance attributable to genetic factors (a2 = 0.52, c2 =0.01, e2 =0.47). Genetic effects on CM were statistically significant (χ2 = 5.43, p-value =0.0098) while shared environmental effects were not significant. Heritability maps of cortical myelination demonstrated substantial regional variability (Figure 1A and Figure S3). Genetic effects were strongest in the right posterior cerebrum, with a clear anterior-posterior gradient. In the right parasagittal occipital lobe, lateral occipital lobe, and parieto-occipital cortex, 60–70% of the phenotypic variance was attributable to genetic factors. Intermediate heritability estimates were seen throughout most of the remainder of the brain. Genetic influences on myelination were statistically significant at the vertex level for the majority of the cortex, with small regions of anterior insula, medial orbitofrontal cortex, and left parieto-occipital cortex not reaching statistical significance. Variance attributable to the shared environment was substantially lower, generally accounting for less than 25% of the phenotypic variance and not statistically significant.
Figure 1:
The genetics of cortical myelination (A), surface area (B), and thickness (C) in the Human Connectome Project. The three top columns represent maximum likelihood estimates for genetic (a2), shared environmental (c2), and unique environmental (e2) variance components. The two lower columns represent FDR-corrected probability maps testing the statistical significance of genetic and shared environmental influences.
There were strong genetic influences on total cerebral surface area, with over 70% of the phenotypic variance attributable to genetic factors (a2 = 0.73, c2=0.00, e2=0.27); genetic effects were statistically significant (χ2 = 43.1, p-value <0.0001). Heritability estimates for SA were generally stronger in the posterior cerebrum, with particularly strong genetic effects observed in the occipital lobes, precuneus, and lateral occipitotemporal cortex (Figure 1B and Figure S4). Genetic effects were statistically significant for the vast majority of the cortex. In contrast, there were negligible influences of the shared environment, with no vertices reaching statistical significance after correction for multiple testing.
Global mean CT was also highly heritable in HCP, with 85% of its variance attributed to additive genetic factors (a2 = 0.85, c2 =0.00, e2 =0.15). Genetic effects were statistically significant (χ2 = 100.1, p-value <0.0001). Regional genetic effects were generally lower in magnitude than either SA or CM and with a distinct spatial distribution (Figure 1C and Figure S5). Regions of highest CT heritability were observed in the posterior frontal and parietal lobes (particularly surrounding the central sulcus), superior temporal lobe, and parasagittal frontal, parietal, and occipital cortex. Genetic effects on thickness were statistically significant for large regions of the cerebral cortex, with superficial cortical vertices generally more significant than those located deep within sulci.
Relationships Between Metrics
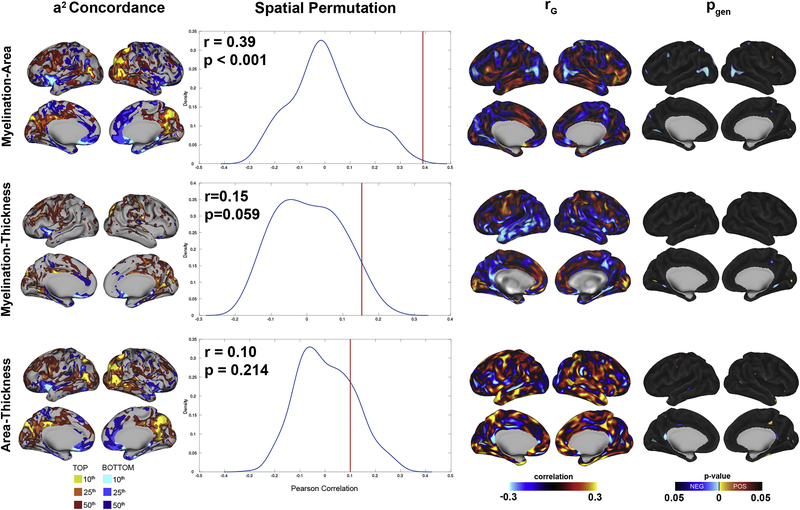
When we examined cortical myelination and surface area simultaneously (Figure 2), there were multiple regions of concordance between CM and SA heritability maps. Both maps had similar hotspots in the parasagittal occipital lobe and precuneus, as well as right occipitotemporal cortex. Genetic influences on the parasagittal frontal lobes and insula were in the lower quartile for both SA and CM. There was statistically significant spatial concordance between CM and SA heritability maps (r = 0.39, p-value <0.001). However, despite similarities in heritability patterns between these measures, genetic correlations between them were relatively low. The highest positive correlations were seen in the bilateral inferior frontal gyri, with the strongest negative correlations in the lateral occipitotemporal cortex. With the exception of the occipitotemporal cortex, most regions’ genetic correlations between SA and CM were not statistically significant. Environmentally-mediated correlations also were not statistically significant at any vertex. Similar findings were seen when examining global relationships between total SA and mean CM, with all correlations approaching zero (rP = 0.03, rG= 0.08, rE= −0.05) and none statistically significant.
Figure 2:
Genetic relationships between all pairwise combinations of neuroanatomic metrics. Concordance maps (left) show brain regions where heritability (a2) centiles are concordant for both metrics. Density plots quantify the observed correlation between heritability maps (red line) overlaid on spatially permuted null distributions. The brain maps on the right present results from bivariate analyses directly testing for shared genetic effects. On the right, genetic correlations (rG) as well as the corresponding FDR-corrected probability maps (pgen) are shown, with the latter color coded based on whether statistically significant correlations were positive (red) or negative (blue).
In contrast to CM-SA relationships, the concordance maps between CM and CT showed very little subjective overlap (Figure 2). Objective comparison via spatial permutation was not statistically significant. Furthermore, there was little evidence of shared genetic influences between CM and CT when they were modeled simultaneously, with only a few vertices reaching statistical significance. Vertex-level genetic correlations demonstrated regional variability, with negative correlations in the temporal lobe and positive correlations in the parasagittal frontal lobe and occipital pole; however, only a few small clusters reached statistical significance. Correlations between global mean CM and CT were weak (rP = −0.05, rG= −0.03, rE= 0.11) and not statistically significant.
For completeness, we also examined the relationships between CT and SA (Figure 2); prior studies have demonstrated that these measures are genetically orthogonal in older adults (Panizzon et al., 2009), but may be correlated in infancy and childhood (Jha et al., 2018; Schmitt et al., 2019a). Consistent with other adult samples, we observed weak global correlations between mean CT and total SA (rP = 0.15, rG= 0.18, rE= 0.01). Although small in magnitude, both genetic (χ2 = 14.7, p-value 0.0001) and phenotypic (χ2 = 19.2, p-value 0.0002) correlations were statistically significant. Vertex-level genetic correlations had a distinct pattern compared to CT-CM and CM-SA, with negative genetic correlations consistently within sulci and positive correlations within gyri. However, genetic effects at the vertex level were generally not statistically significant after correction for multiple testing.
Correlates With Intelligence
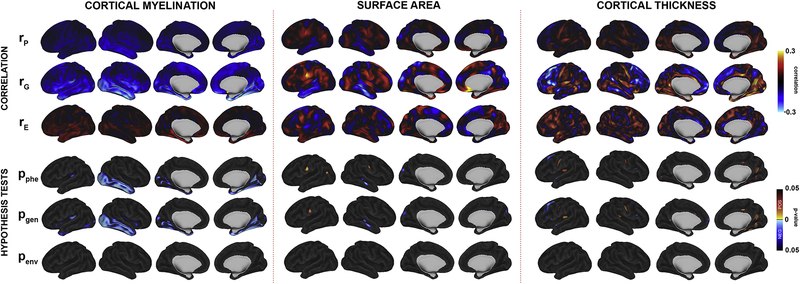
Associations between NTC and all neuroanatomic metrics are summarized in Figure 3 and supplementary figures S6–S8. There were regional but relatively weak negative phenotypic and genetic correlations between CM and NTC, primarily localized to the right greater than left insula and inferior occipitotemporal cortex. Genetically-mediated associations were statistically significant, with probability maps having a similar pattern to genetic correlation maps. NTC-SA correlations were more variable, with the strongest positive genetic correlations in the left perisylvian frontal and parietal cortex. There were a few small clusters that were significant after correction for multiple testing. There were significant positive genetically-mediated NTC-CT associations in the inferior occipitotemporal cortex and positive associations in premotor cortex and cingulate, although the magnitude of these effects was relatively weak. Environmentally mediated relationships between intelligence and brain measures were not statistically significant.
Figure 3:
Results from bivariate models testing genetically-mediated associations between a measure of general intelligence (NTC) with CM (left), SA (center), and CT (right). Genetic (rG), environmental (rE), and phenotypic (rP) correlations are provided, as well as corresponding FDR-corrected probability maps color coded based on whether statistically significant correlations were positive (red) or negative (blue).
Discussion
Similar to other cortical phenotypes, we observed a high degree of regional variability in the heritability of CM; the highest estimates were observed in the posterior cerebrum. To our knowledge, the current study is the first to report heritability estimates of CM at the vertex level. The overall patterns that we observed were similar to that of the only other genetically-informative study on CM in the literature (Liu et al., 2018). Similarities between the two analyses were expected since both use HCP data. The studies differ in that Liu et al. was based on data from the earlier S900 HCP release (N=873), used 210 regions of interest (ROI) rather than vertex-level measures, employed the SOLAR software package rather than OpenMx for quantitative genetic modeling, and presented heritability maps based on AE rather than ACE models (although ACE model parameter estimates were provided as supplementary data). ROI parcellation versus vertex-level measures may explain small differences in the magnitude of the observed genetic effects, as measurement error with ROI analyses improves at the cost of reduced spatial resolution. Sampling variation may also contribute to discrepancies. Nevertheless, it is reassuring to observe strong similarities between the two studies despite their methodological differences.
Neurodevelopmental trajectories in cortical myelination in humans are distinct when compared to nonhuman primates, with prolonged myelination in our species extending well into the third decade of life (Miller et al., 2012). The more recently evolutionarily expanded regions of the human cortex tend to be more lightly myelinated compared to nonhuman primates; higher order cortex is therefore also generally less myelinated relative to primary cortex (Glasser et al., 2014). Although some regions with the highest heritability corresponded to regions of greatest myelination (e.g. cuneus), other highly-myelinated regions had relatively low genetically mediated variation (e.g. primary motor cortex). In general, genetic factors had the strongest role on individual differences in regions of the cerebrum involved in visual processing, although it is noteworthy that there was evidence of at least modest genetic effects throughout most of the cerebral cortex.
As hypothesized, we found that genetic effects on SA were strongest in the posterior cerebrum, particularly in the occipital lobe. This finding is concordant with multiple prior studies on the genetics of cerebral surface area, particularly to those on childhood and younger adult samples (Jha et al., 2018; Schmitt et al., 2019a; Yoon et al., 2012). Heritability estimates overall were somewhat higher than those observed in younger samples, with stronger genetic effects in the anterior cerebrum. Patterns were similar to those seen Genetics of Brain Structure (GOBS) adult sample (McKay et al., 2014) but were overall lower than those from the Vietnam Era Twin Study of Aging (VETSA) whose mean age was 55.8 (Eyler et al., 2012). Considered in aggregate, these findings suggest that genetic and environmental variation in SA changes over the life cycle.
Patterns of regional heritability CT were also similar to prior studies on younger adult populations, with expected higher regional genetic effects in the parietal lobe and posterior frontal lobe (Joshi et al., 2011; Shen et al., 2016). Estimates were somewhat lower in magnitude compared to our prior estimates of CT heritability on a different subsample from HCP, although patterns were similar (Schmitt et al., 2019b). To facilitate cross-metric comparisons, the current CT data were processed with MSM registration (Robinson et al., 2014), while previously we used standard FreeSurfer vertex-level CT measurements. FreeSurfer registration is based on geometric features such as sulcal curvature and depth, while MSM incorporates additional information on myelination and functional connectivity. Given that individual differences in brain geometry may explain these discrepancies, future studies on the genetics of regional brain morphology may prove interesting.
Cortical Myelination, Surface Area, and Thickness Are Largely Genetically Independent in Adults
In contrast to the strong genetic influences we observed in our univariate analyses, we found very little evidence of shared genetic influences between any of our neuroanatomic measures, either globally or at high spatial resolution. The weak genetic correlation that we observed between global SA and CT (rG=0.18) was not surprising, as it is similar to estimates observed in the GOBS (rG=−0.15), VETSA (rG=0.08), and QITM (rG=−0.21) samples (Panizzon et al., 2009; Strike et al., 2018; Winkler et al., 2010).
However, given the observed similarities between CM and SA heritability maps as well evidence of CM-SA neurodevelopmental concordance (Cafiero et al., 2019), we expected to find stronger shared genetic effects between these two metrics. Surface area expansion is thought to be influenced by numerous factors including intracortical myelination (Fjell et al., 2015); however we found no evidence that these variables are genetically coupled, at least in adults. We similarly expected to observe stronger shared genetic factors between CT and CM, since there is evidence of CT-CM neurodevelopmental coupling in adolescence (Whitaker et al., 2016). Given that genetic associations between surface area and cortical thickness vary with age (Schmitt et al., 2019a), stronger shared genetic effects between these measures may be present in younger subjects; further research on CM-SA and CM-CT relationships at other stages of the life cycle may be of value.
Relationships with Intelligence
Finally, we examined the associations between our neuroanatomic measures and cognitive function. Although prior studies have reported genetically-mediated associations between IQ and white matter fiber integrity (Chiang et al., 2009), to our knowledge the current study is the first to examine cortical myelination specifically. Genome-wide association studies have found evidence that many genes involved in myelination are also associated with intelligence (Hill et al., 2018). We found weak but statistically significant genetically-mediated correlations between NTC and myelination in the inferior temporal and insular cortex. Correlations were negative, indicating that decreases in CM in this region associated with increases in cognitive function. Given that myelin content is associated with increased processing speeds, an inverse relationship may seem counterintuitive. However, intracortical circuit complexity is also inversely correlated with myelination, and myelination has been hypothesized to inhibit neuronal plasticity (Glasser et al., 2014). Furthermore, most higher cortical regions in the brain tend to be more lightly myelinated (Glasser and Van Essen, 2011). Although the literature on the associations between cognition and cortical myelination is sparse, Grydeland et al. found that in adults, a metric of performance variability was most correlated to myelination in the right insula, superior temporal gyrus, and left lingual gyrus (Grydeland et al., 2013).
Genetically-mediated associations between NTC and SA were weaker and largely non-significant, although they generally paralleled our observations that strongest effects are greater in the left than the right perisylvian cortex (Schmitt et al., 2019a). Similarly, correlational patterns between NTC and CT were similar in pattern to correlations between CT and intelligence quotient (IQ) in late adolescence, although much smaller in magnitude (Schmitt et al., 2019b). Given that our prior findings were in a pediatric sample, the observed differences in the strength of correlations may be age related and warrant further investigation.
Supplementary Material
Figure S1: The genetics of cortical myelination (A), surface area (B), and thickness (C) in the Human Connectome Project analyzed with an ACET extended twin design model. The four top columns represent maximum likelihood estimates for genetic (a2), shared environmental (c2), unique environmental (e2), and twin-specific environmental (t2) variance components. The three lower columns represent FDR-corrected probability maps testing the statistical significance of genetic, shared environmental, and twins-pecific environmental influences.
Figure S2: The genetics of cortical myelination (A), surface area (B), and thickness (C) in for all twin-pairs in the S1200 release of the Human Connectome Project (188 MZ and 298 DZ). The three top columns represent maximum likelihood estimates for genetic (a2), shared environmental (c2), and unique environmental (e2) variance components. The two lower columns represent FDR-corrected probability maps testing the statistical significance of genetic and shared environmental influences.
Figure S3: Magnified MLE and probability maps for cortical myelination analyzed with an extended-twin design (same results as Figure 1A).
Figure S4: Magnified MLE and probability maps for surface area analyzed with an extended-twin design (same results as Figure 1B).
Figure S5: Magnified MLE and probability maps for cortical myelination analyzed with an extended-twin design (same results as Figure 1C).
Figure S6: Magnified MLE and probability maps from bivariate models testing genetically-mediated associations between a NTC and CM.
Figure S7: Magnified MLE and probability maps from bivariate models testing genetically-mediated associations between a NTC and SA.
Figure S8: Magnified MLE and probability maps from bivariate models testing genetically-mediated associations between a NTC and CT.
Acknowledgements
This work was supported by National Institute of Mental Health grant MH-20030 and Big Data to Knowledge (BD2K) grant K01-ES026840. AR and SL were supported by the intramural program of the National Institutes of Health (Clinical trial NCT00001246. clinicaltrials.gov; NIH Annual Report Number. ZIA MH002794-13). Data were provided by the Human Connectome Project. WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander-Bloch AF, Shou H, Liu S, Satterthwaite TD, Glahn DC, Shinohara RT, Vandekar SN, Raznahan A, 2018. On testing for spatial correspondence between maps of human brain structure and function. Neuroimage 178, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad M, Stanley JA, Raz N, 2017. Test-retest reliability and concurrent validity of in vivo myelin content indices: Myelin water fraction and calibrated T1w/T2w image ratio. Hum. Brain Mapp 38, 1780–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boker S, Neale M, Maes H, Wilde M, Spiegel M, 2011. OpenMx: an open source extended structural equation modeling framework. Psychometrika 76, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafiero R, Brauer J, Anwander A, Friederici AD, 2019. The Concurrence of Cortical Surface Area Expansion and White Matter Myelination in Human Brain Development. Cereb. cortex 29, 827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang M-C, Barysheva M, Shattuck DW, Lee AD, Madsen SK, Avedissian C, Klunder AD, Toga AW, McMahon KL, de Zubicaray GI, Wright MJ, Srivastava A, Balov N, Thompson PM, 2009. Genetics of Brain Fiber Architecture and Intellectual Performance. J. Neurosci 29, 2212–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominicus A, Skrondal A, Gjessing HK, Pedersen NL, Palmgren J, 2006. Likelihood ratio tests in behavioral genetics: problems and solutions. Behav. Genet 36, 331–40. [DOI] [PubMed] [Google Scholar]
- Edwards A, 1972. Likelihood: an account of the statistical concept of likelihood and its application to scientific inference. University Press, Cambridge. [Google Scholar]
- Eyler LT, Chen C-H, Panizzon MS, Fennema-Notestine C, Neale MC, Jak A, Jernigan TL, Fischl B, Franz CE, Lyons MJ, Grant M, Prom-Wormley E, Seidman LJ, Tsuang MT, Fiecas MJA, Dale AM, Kremen WS, 2012a. A comparison of heritability maps of cortical surface area and thickness and the influence of adjustment for whole brain measures: a magnetic resonance imaging twin study. Twin Res. Hum. Genet 15, 304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyler LT, Chen C-H, Panizzon MS, Fennema-Notestine C, Neale MC, Jak A, Jernigan TL, Fischl B, Franz CE, Lyons MJ, Grant M, Prom-Wormley E, Seidman LJ, Tsuang MT, Fiecas M.J. a, Dale AM, Kremen WS, 2012b. A comparison of heritability maps of cortical surface area and thickness and the influence of adjustment for whole brain measures: a magnetic resonance imaging twin study. Twin Res. Hum. Genet 15, 304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, 2012. FreeSurfer. Neuroimage 62, 774–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell AM, Westlye LT, Amlien I, Tamnes CK, Grydeland H, Engvig A, Espeseth T, Reinvang I, Lundervold AJ, Lundervold A, Walhovd KB, 2015. High-expanding cortical regions in human development and evolution are related to higher intellectual abilities. Cereb. Cortex 25, 26–34. [DOI] [PubMed] [Google Scholar]
- Ganzetti M, Wenderoth N, Mantini D, 2014. Whole brain myelin mapping using T1- and T2-weighted MR imaging data. Front. Hum. Neurosci 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T, 2002. Thresholding of Statistical Maps in Functional Neuroimaging Using the False Discovery Rate. Neuroimage 15, 870–878. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Goyal MS, Preuss TM, Raichle ME, Van Essen DC, 2014. Trends and properties of human cerebral cortex: Correlations with cortical myelin content. Neuroimage 93, 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M, 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC, 2011. Mapping Human Cortical Areas In Vivo Based on Myelin Content as Revealed by T1- and T2-Weighted MRI. J. Neurosci 31, 11597–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grydeland H, Walhovd KB, Tamnes CK, Westlye LT, Fjell AM, 2013. Intracortical Myelin Links with Performance Variability across the Human Lifespan: Results from T1- and T2-Weighted MRI Myelin Mapping and Diffusion Tensor Imaging. J. Neurosci 33, 18618–18630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton RK, Akshoomoff N, Tulsky D, Mungas D, Weintraub S, Dikmen S, Beaumont J, Casaletto KB, Conway K, Slotkin J, Gershon R, 2014. Reliability and validity of composite scores from the NIH toolbox cognition battery in adults. J. Int. Neuropsychol. Soc 20, 588–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, Inder T, Neil J, Dierker D, Harwell J, Van Essen D, 2010. Similar patterns of cortical expansion during human development and evolution. Proc. Natl. Acad. Sci 107, 13135–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WD, Marioni RE, Ritchie OMSJ, Hagenaars SP, Mcintosh AM, Davies G, Deary IJ, 2018. A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Xia K, Schmitt JE, Ahn M, Girault J, Murphy V, Li G, Wang L, Shen D, Zou F, Zhu H, Styner M, Knickmeyer R, Gilmore J, 2018. Genetic Influences on Neonatal Cortical Thickness and Surface Area. Hum. Brain Mapp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AA, Leporé N, Joshi SH, Lee AD, Barysheva M, Stein JL, McMahon KL, Johnson K, De Zubicaray GI, Martin NG, Wright MJ, Toga AW, Thompson PM, 2011. The contribution of genes to cortical thickness and volume. Neuroreport 22, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Schmitt JE, Ordaz SJ, Wallace GL, Neale MC, Lerch JP, Kendler KS, Evans AC, Giedd JN, 2009. Differences in genetic and environmental influences on the human cerebral cortex associated with development during childhood and adolescence. Hum. Brain Mapp 30, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li A, Zhu M, Li J, 2018. Genetic influences on cortical myelination in the human brain. Genes. Brain. Behav in press. [DOI] [PubMed] [Google Scholar]
- Marcus DS, Harms MP, Snyder AZ, Jenkinson M, Wilson JA, Glasser MF, Barch DM, Archie KA, Burgess GC, Ramaratnam M, Hodge M, Horton W, Herrick R, Olsen T, McKay M, House M, Hileman M, Reid E, Harwell J, Coalson T, Schindler J, Elam JS, Curtiss SW, Van Essen DC, 2013. Human Connectome Project informatics: Quality control, database services, and data visualization. Neuroimage 80, 202– [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DR, Knowles EEM, Winkler AAM, Sprooten E, Kochunov P, Olvera RL, Curran JE, Kent JW, Carless MA, Goring HHH, Dyer TD, Duggirala R, Almasy L, Fox PT, Blangero J, Glahn DC, 2014. Influence of age, sex and genetic factors on the human brain. Brain Imaging Behav. 8, 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, Mcarthur MJ, 2012. Prolonged myelination in human neocortical evolution. Proc. Natl. Acad. Sci 109, 16480–16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M, Cardon L, 1992. Methodology for Genetic Studies of Twins and Families. Kluver, Dordrecht, The Netherlands. [Google Scholar]
- Neale MC, Hunter MD, Pritikin JN, Zahery M, Brick TR, Kirkpatrick RM, Estabrook R, Bates TC, Maes HH, Boker SM, 2016. OpenMx 2.0: Extended Structural Equation and Statistical Modeling. Psychometrika 81, 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizzon MS, Fennema-Notestine C, Eyler LT, Jernigan TL, Prom-Wormley E, Neale M, Jacobson K, Lyons MJ, Grant MD, Franz CE, Xian H, Tsuang M, Fischl B, Seidman L, Dale A, Kremen WS, 2009. Distinct genetic influences on cortical surface area and cortical thickness. Cereb. Cortex 19, 2728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D, Boomsma DI, 2000. A note on the statistical power in extended twin designs. Behav. Genet 30, 147–58. [DOI] [PubMed] [Google Scholar]
- Rimol LM, Panizzon MS, Fennema-Notestine C, Eyler LT, Fischl B, Franz CE, Hagler DJ, Lyons MJ, Neale MC, Pacheco J, Perry ME, Schmitt JE, Grant MD, Seidman LJ, Thermenos HW, Tsuang MT, Eisen SA, Kremen WS, Dale AM, 2010. Cortical Thickness Is Influenced by Regionally Specific Genetic Factors. Biol. Psychiatry 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Smith SM, Van Essen DC, Jenkinson M, 2014. MSM: A new flexible framework for multimodal surface matching. Neuroimage 100, 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Neale MC, Clasen LS, Liu S, Seidlitz J, Pritikin JN, Wallace GL, Lee NR, Giedd JN, Raznahan A, 2019a. A Comprehensive Quantitative Genetic Analysis of Cerebral Surface Area in Youth. J. Neurosci 39, 3028–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Neale MC, Fassassi B, Perez J, Lenroot RK, Wells EM, Giedd JN, 2014. The dynamic role of genetics on cortical patterning during childhood and adolescence. Proc. Natl. Acad. Sci. U. S. A 111, 6774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JE, Raznahan A, Clasen LS, Wallace GL, Pritikin JN, Lee NR, Giedd JN, Neale MC, 2019b. The Dynamic Associations Between Cortical Thickness and General Intelligence are Genetically Mediated. Cereb. Cortex 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafee R, Buckner RL, Fischl B, 2015. Gray matter myelination of 1555 human brains using partial volume corrected MRI images. Neuroimage 105, 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen KK, Doré V, Rose S, Fripp J, Mcmahon KL, de Zubicaray GI, Martin NG, Thompson PM, Wright MJ, Salvado O, 2016. Heritability and genetic correlation between the cerebral cortex and associated white matter connections. Hum. Brain Mapp. 37, 2331–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strike LT, Hansell NK, Couvy-Duchesne B, Thompson PM, de Zubicaray GI, McMahon KL, Wright MJ, 2018. Genetic Complexity of Cortical Structure: Differences in Genetic and Environmental Factors Influencing Cortical Surface Area and Thickness. Cereb. Cortex 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2018. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Uddin MN, Figley TD, Marrie RA, Figley CR, 2018. Can T1w/T2w ratio be used as a myelin-specific measure in subcortical structures? Comparisons between FSE-based T1w/T2w ratios, GRASE-based T1w/T2w ratios and multi-echo GRASE-based myelin water fractions. NMR Biomed. 31, 1–11. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E, 2012. The Human Connectome Project: A data acquisition perspective. Neuroimage 62, 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandekar SN, Shinohara RT, Raznahan A, Roalf DR, Ross M, DeLeo N, Ruparel K, Verma R, Wolf DH, Gur RC, Gur RE, Satterthwaite TD, 2015. Topologically Dissociable Patterns of Development of the Human Cerebral Cortex. J. Neurosci 35, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker KJ, Vértes PE, Romero-Garcia R, Váša F, Moutoussis M, Prabhu G, Weiskopf N, Callaghan MF, Wagstyl K, Rittman T, Tait R, Ooi C, Suckling J, Inkster B, Fonagy P, Dolan RJ, Jones PB, Goodyer IM, Bullmore ET, 2016. Adolescence is associated with genomically patterned consolidation of the hubs of the human brain connectome. Proc. Natl. Acad. Sci 113, 9105–9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford TJ, Ford JM, Mathalon DH, Kubicki M, Shenton ME, 2012. Schizophrenia, myelination, and delayed corollary discharges: A hypothesis. Schizophr. Bull 38, 486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, Duggirala R, Glahn DC, 2010. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage 53, 1135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon U, Perusse D, Evans AC, 2012. Mapping genetic and environmental influences on cortical surface area of pediatric twins. Neuroscience 220, 169–178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: The genetics of cortical myelination (A), surface area (B), and thickness (C) in the Human Connectome Project analyzed with an ACET extended twin design model. The four top columns represent maximum likelihood estimates for genetic (a2), shared environmental (c2), unique environmental (e2), and twin-specific environmental (t2) variance components. The three lower columns represent FDR-corrected probability maps testing the statistical significance of genetic, shared environmental, and twins-pecific environmental influences.
Figure S2: The genetics of cortical myelination (A), surface area (B), and thickness (C) in for all twin-pairs in the S1200 release of the Human Connectome Project (188 MZ and 298 DZ). The three top columns represent maximum likelihood estimates for genetic (a2), shared environmental (c2), and unique environmental (e2) variance components. The two lower columns represent FDR-corrected probability maps testing the statistical significance of genetic and shared environmental influences.
Figure S3: Magnified MLE and probability maps for cortical myelination analyzed with an extended-twin design (same results as Figure 1A).
Figure S4: Magnified MLE and probability maps for surface area analyzed with an extended-twin design (same results as Figure 1B).
Figure S5: Magnified MLE and probability maps for cortical myelination analyzed with an extended-twin design (same results as Figure 1C).
Figure S6: Magnified MLE and probability maps from bivariate models testing genetically-mediated associations between a NTC and CM.
Figure S7: Magnified MLE and probability maps from bivariate models testing genetically-mediated associations between a NTC and SA.
Figure S8: Magnified MLE and probability maps from bivariate models testing genetically-mediated associations between a NTC and CT.