Figure 2.
Structural overview of the SARS-CoV-2 RNA genome
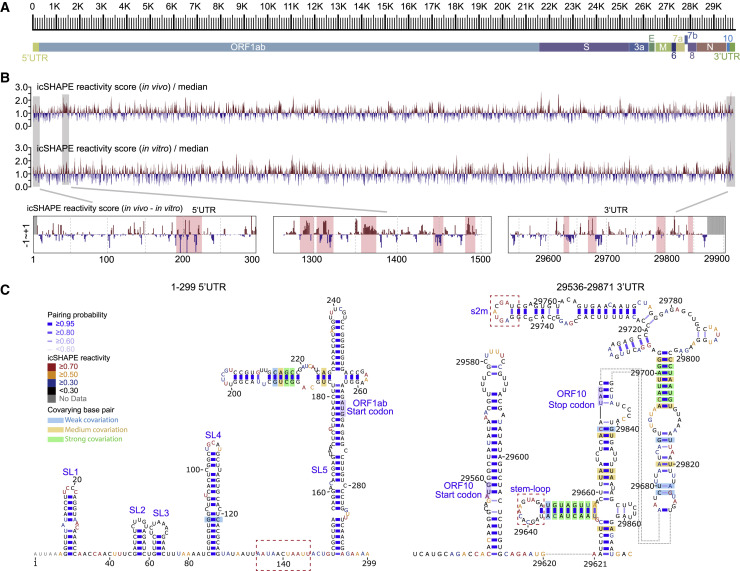
(A) Scale marker for the 30-kb SARS-CoV-2 RNA genome (top) and a genic model showing the known organization of the genome into the 5′ UTR, the two known ORFs (ORF1a and ORF1b), nine major subgenomic RNAs, and the 3′ UTR (bottom).
(B) Top track: Normalized icSHAPE reactivity scores across the whole SARS-CoV-2 genome based on in vivo data, shown relative to the global median value, with higher values corresponding to more flexible nucleotides. Blue represents a nucleotide more likely to be base paired, whereas red represents a nucleotide more likely to be single-stranded. The normalized scores have been smoothed using a 30-nt window size. Middle track: Normalized genome-wide icSHAPE reactivity scores for the SARS-CoV-2 genome based on the in vitro refolding data. Bottom tracks: in vivo and in vitro icSHAPE reactivity score differences (in vivo – in vitro) of the 5′UTR, an example ORF1ab region, and the 3′UTR. Significant regions are highlighted with red boxes.
(C) RNA structure models of the SARS-CoV-2 5′-UTR (left) and 3′-UTR (right) (both with flanking regions), constructed with the RNAstructure program using the icSHAPE reactivity scores as constraints. Nucleotides are colored by icSHAPE reactivity scores, with red and yellow colors indicating reactive nucleotides. Blue bars show the probability of base pairing. Nucleotides with a color background were predicted as co-variant base pairs. The red dashed boxes label the structural regions with differences in comparison with Rangan’s structural models (Rangan et al., 2020).
See also Figures S1 and S2; Tables S1 and S2.