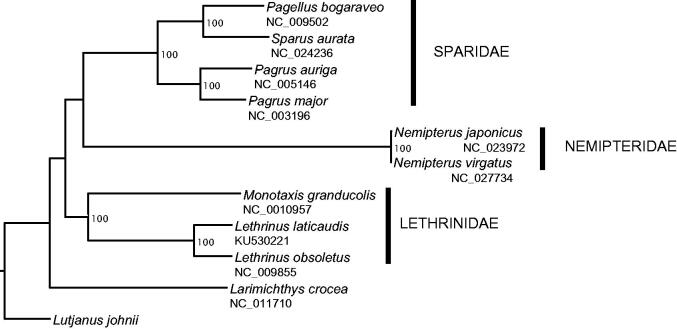
Figure 1.
Phylogenetic tree of 11 closely related species including Lethrinus laticaudis based on the analysis of mitogenome sequences. The mitogenomes were aligned in Geneious using ClustalW alignment method with default settings. Poorly aligned positions and indels were removed with Gblock v 0.91b (Castresana 2000; Dereeper et al. 2008) using default settings and the D-Loop region was also excluded (total length: 15,323 bp). A heuristic maximum likelihood (ML) search was conducted using RaxML HPC v8 (Stamatakis 2006) on XSEDE, implemented in the CyberInfrastructure for Phylogenetic Research (CIPRES) portal v3.3 (http://www.phylo.org/portal2, Miller et al. 2010). Lutjanus johnii was set as the outgroup species for our analysis. A rapid bootstrap analysis and a search for best-scoring ML tree were performed. Robustness of the nodes was assessed with 1000 bootstrap replicates.