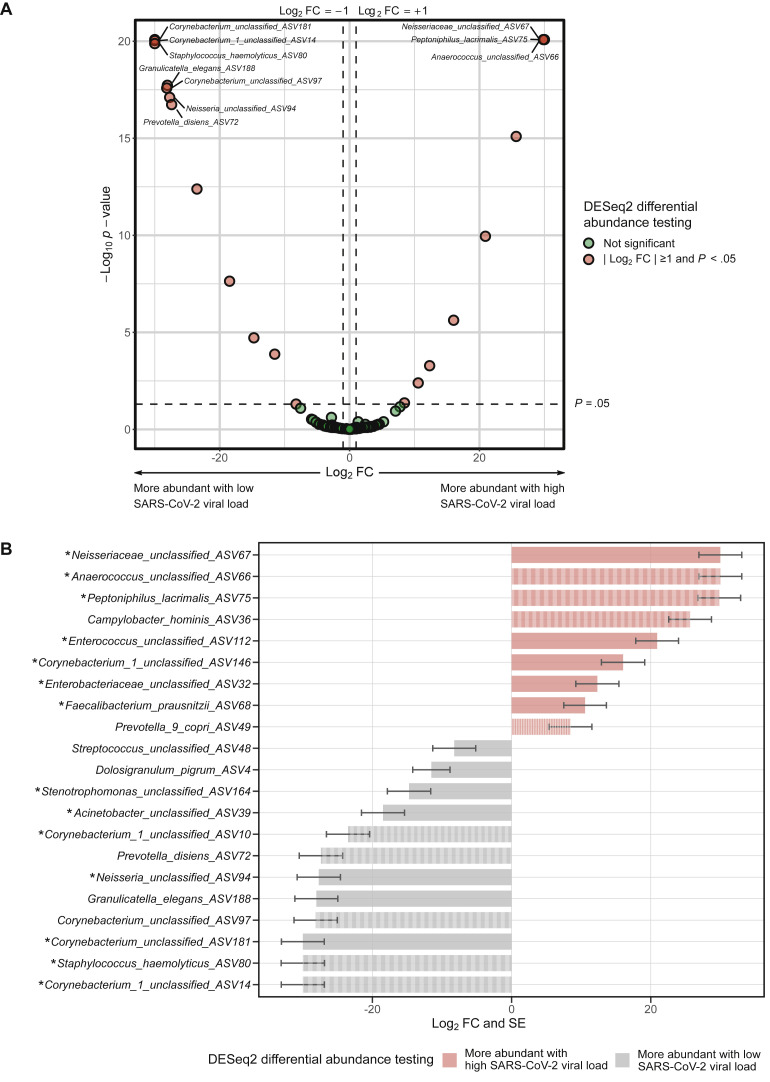
Fig 4.
Differences in the abundance of taxa of the URT microbiome between SARS-CoV-2–infected adults with and without high viral load (defined as a quantitative reverse transcription PCR cycle threshold value below the median for the detection of SARS-CoV-2 nucleocapside gene region 1 [N1]). Differential abundance testing was conducted using DESeq2 models at the ASV level including age and sex as covariates. A, Volcano plot of log2 fold change (FC) vs statistical significance. The red circles indicate ASVs that were significantly different between groups. Only the top 10 most significantly different ASVs are labeled. B, Bar plot depicting the log2 FCs and SEs for ASVs that were significantly different between groups. The asterisks indicate ASVs that were significantly different between groups and had a consistent direction of association in similar DESeq2 analyses that used a definition of high viral load based on a quantitative reverse transcription PCR cycle threshold value below the median for the detection of SARS-CoV-2 nucleocapside gene region 2 (N2). The striped bars indicate ASVs that were significantly different between groups and had a consistent direction of association in similar DESeq2 analyses comparing adults with and without SARS-CoV-2 infection.