Extended Data Figure 1. Additional characterization of progeria patient-derived cells treated with ABE7.10max-VRQR.
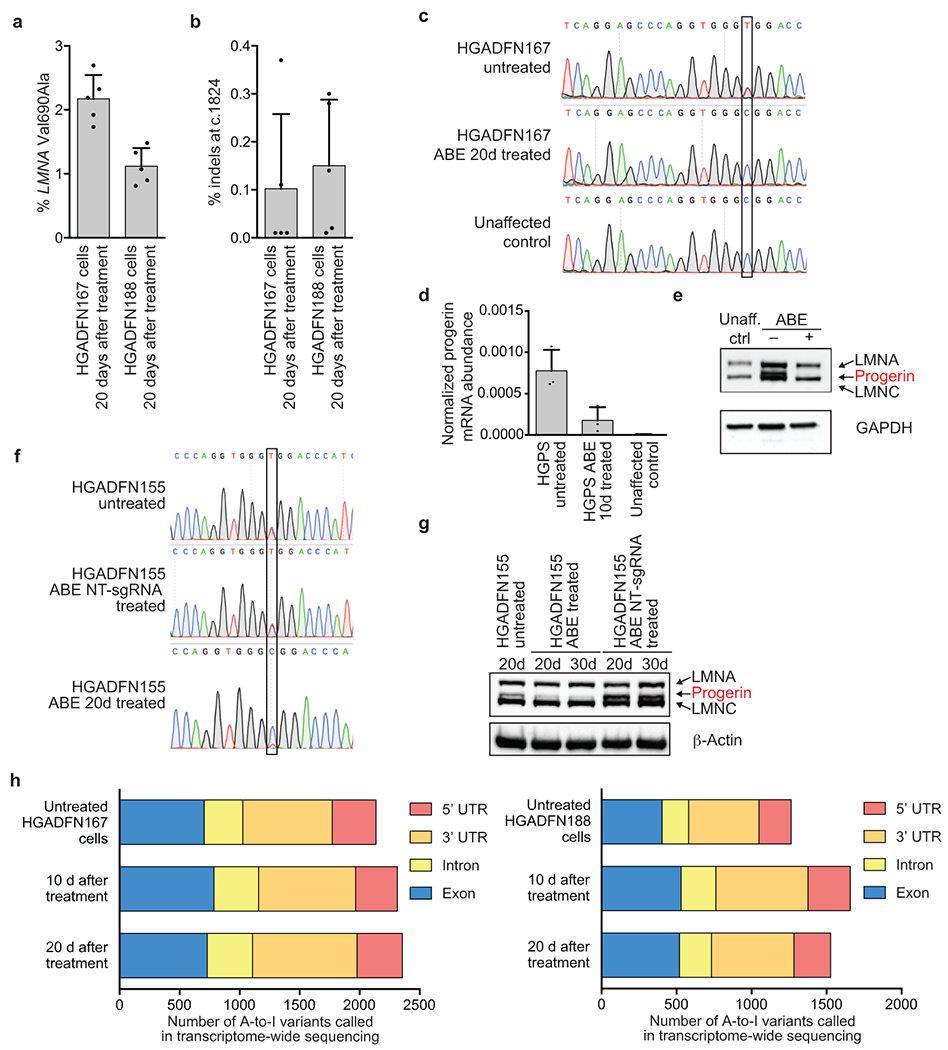
(a) Bystander Val690Ala editing in progeria patient-derived HGADFN167 and HGADFN188 cells 20 days after treatment with lentiviral ABE7.10max-VRQR. (b) Indel formation frequency at the c.1824 target locus in HGADFN167 and HGADFN188 cells 20 days after treatment with lentiviral ABE7.10max-VRQR. Values and error bars represent mean±SD for n=5 technical replicates (individual points) for (a) and (b). (c) Sanger DNA sequencing traces of untreated HGADFN167 cells, 20-day treated HGADFN167 cells, and unaffected control cells. The target nucleotide is boxed. (d) qPCR-normalized progerin mRNA abundance in cells described in c. Values and error bars represent mean±SD for n=3 biological replicates. (e) Western blot analysis of HGADFN167 cells described in c. LMNA, progerin, and LMNC protein are all stained on the gel, A GAPDH loading control is shown below. An additional replicate is provided in Fig. 1d. Unaff. ctrl, control cells from an unaffected parent. (f) Sanger DNA sequencing traces of untreated, non-targeting (NT) sgRNA treated, and ABE treated HGADFN155 fibroblasts at 20d and 30d time-points to ensure NT-sgRNA did not lead to DNA editing. (g) Western blot analysis of cells described in f. LMNA, progerin, and LMNC proteins are all stained on the gel, A β-actin loading control is shown below. Expected molecular weights: lamin A, 74 kDa; progerin, 69 kDa; lamin C, 65 kDa. Complete blots are available in Supplementary Figure 1. Additional replication was not performed. (h) HGADFN167 (left) and HGADFN188 (right) cell lines untreated or treated with lentiviral ABE7.10max-VRQR after 10 or 20 days show similar relative distributions of A-to-I single-nucleotide variants (SNVs) in their transcriptomes compared with the hg38 human genome reference sequence. On average, 36±3.6% of SNPs in these samples occur with ~100% frequency, suggesting they arise from genomic sequence variations; however, we cannot explicitly exclude them from consideration since no whole genome sequence is available for these cell lines. Raw counts of 100% edited SNPs per sample are: Untreated HGADFN167 cells (849), HGADFN167 10 d after treatment (883), HGADFN167 20 d after treatment (871), Untreated HGADFN188 cells (488), HGADFN188 10 d after treatment (501), HGADFN188 20 d after treatment (510).
