Figure 3. Molecular modelling of identified O-glycosites.
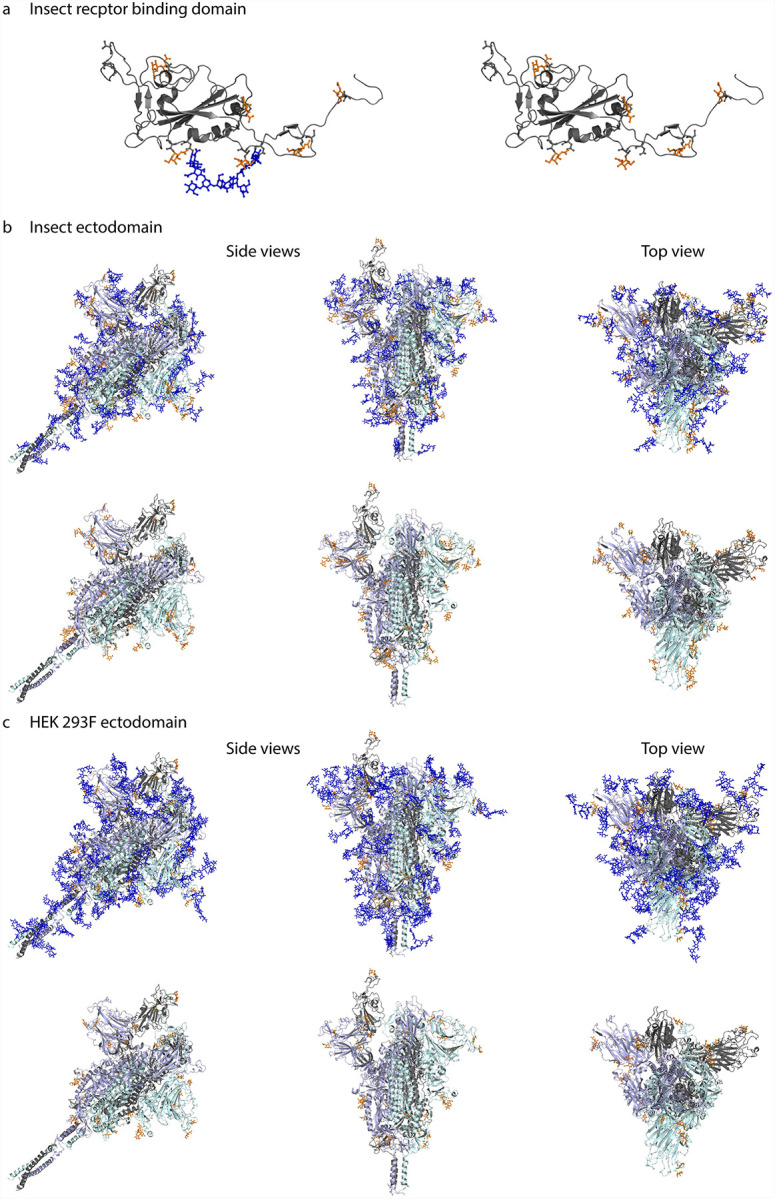
Identified site-specific O-glycans (orange) and known N-glycans (blue) were modelled onto 3D reconstructions of SARS-CoV-2 S protein. Sites found on monomeric RBD expressed in insect cells (a), ectdomain expressed in insect cells (b), or ectodomain expressed in HEK 293F cells (c) are shown. Longest identified O-glycan structures are presented at specific sites of each protein. Paucimannose type N-glycan structures are shown on the insect cell-expressed products, whereas structures defined in the full-length glycosylated model by Woo et al. [28] are shown on the HEK 293F-expressed ectodomains. Models with and without N-glycans are included in each panel. Only the amino acids covered by different construct designs are shown.
