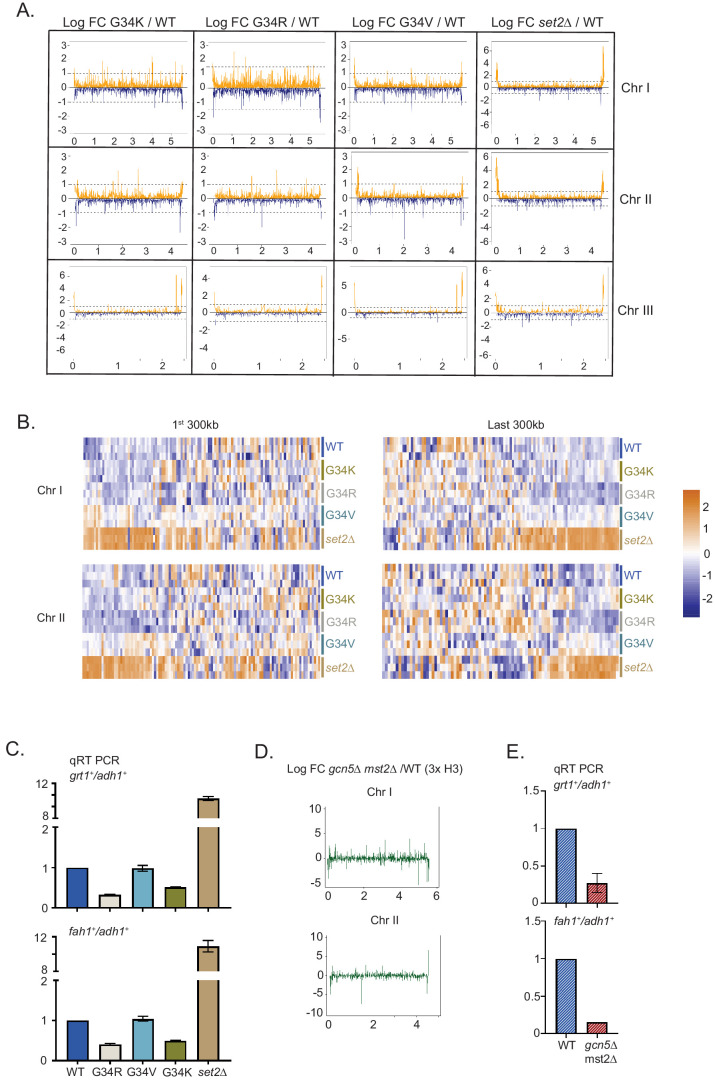
Figure 4. Distinct transcriptional outcomes for H3-G34 mutants: substitution with basic residues suppressing some subtelomeric transcripts, as seen in strains deficient in H3 K36 acetylation.
(A) RNA-seq profiles for chromosomes I, II, and III comparing Logfold change ratios for H3-G34K/H3-WT, H3-G34V/H3-WT, H3-G34R/H3-WT, or set2Δ/H3-WT plotted against chromosome coordinates. (B) Zoomed-in regions of Chr I (first 300 Kb and last 300 Kb, top) and Chr II (first 300 Kb and last 300 Kb, bottom) showing Z scores of log2 CPM for individual biological replicates. (C) qRT-PCR validation of ST genes fah1+ and grt1+ expression relative to adh1+ expression from two independent biological replicates. Samples were normalized to the WT-H3 strain. Subtelomeric transcripts in H3-G34R and H3-G34K are repressed compared with H3-WT and upregulated in set2Δ. (D) Chromosome-wide plots of transcriptional regulation in gcn5Δmst2Δ cells (3xH3) compared with wild type for Chr I and Chr II. Data reanalyzed from Nugent et al., 2010. (E) qRT-PCR validation of fah1+ and grt1+ expression relative to adh1+ expression from two independent biological replicates. Samples were normalized to the WT-H3 strain. Subtelomeric transcripts in gcn5Δmst2Δ cells (3xH3) are reduced compared with wild type.