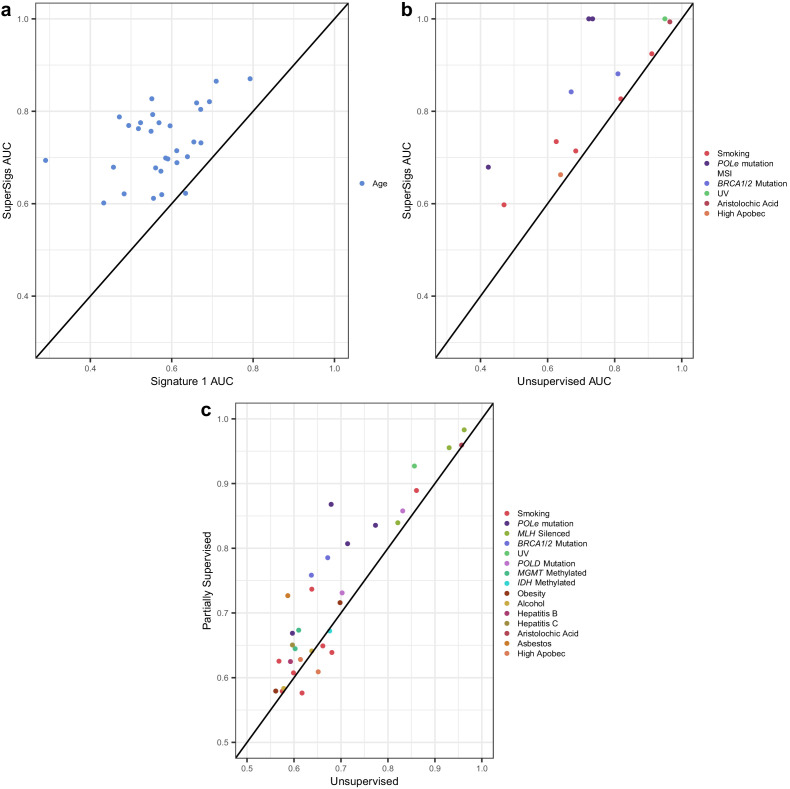
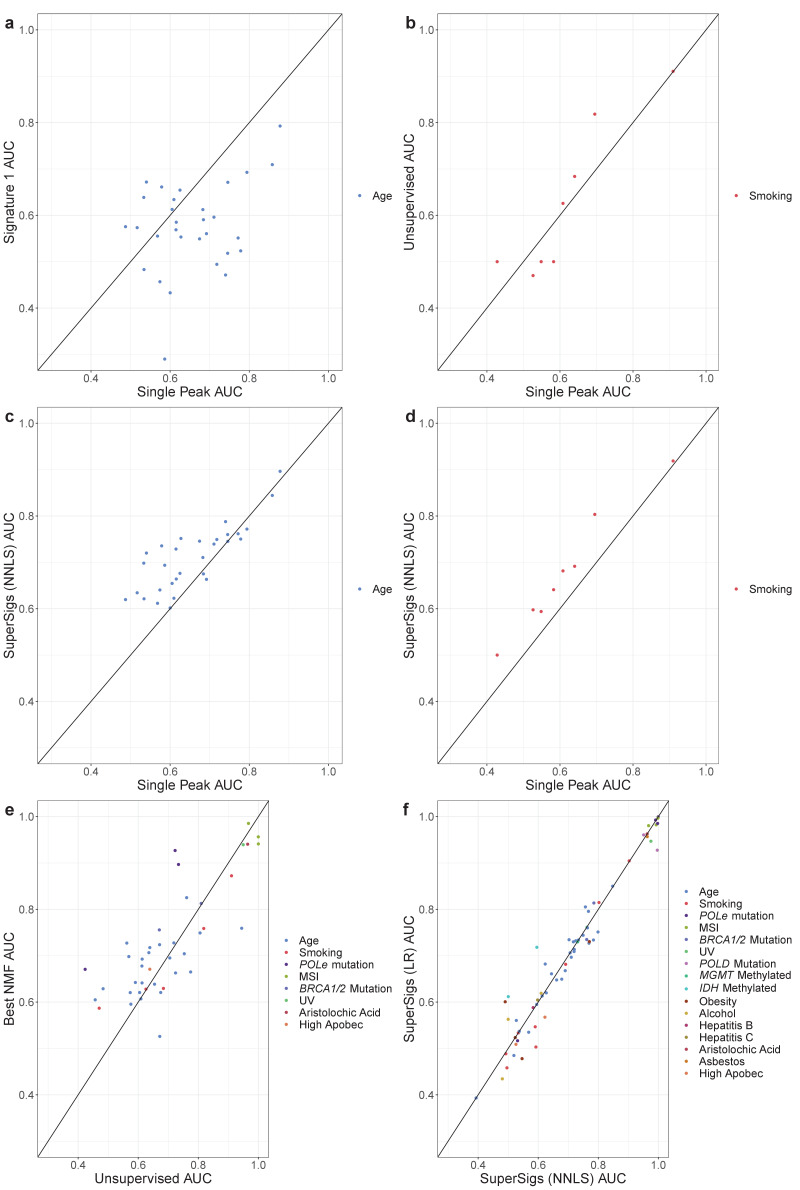
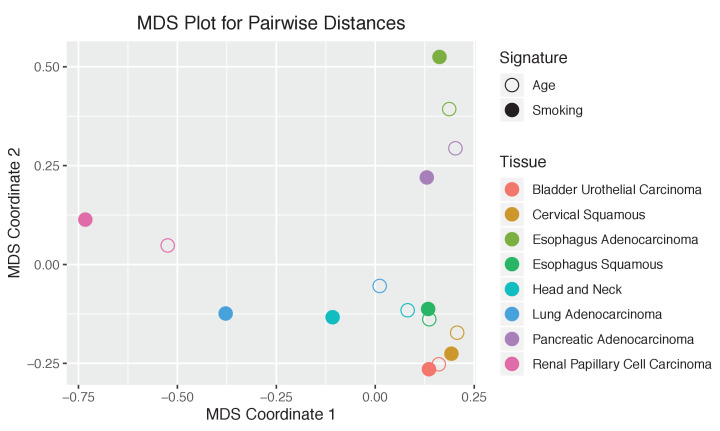
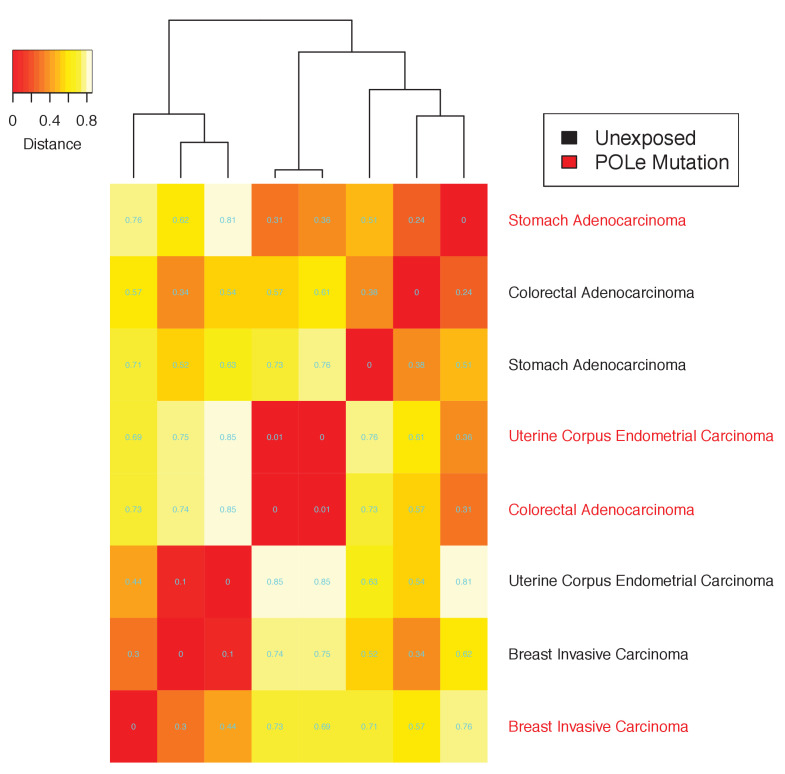
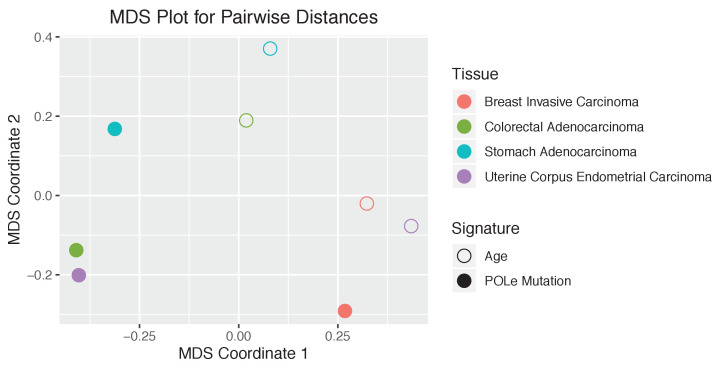
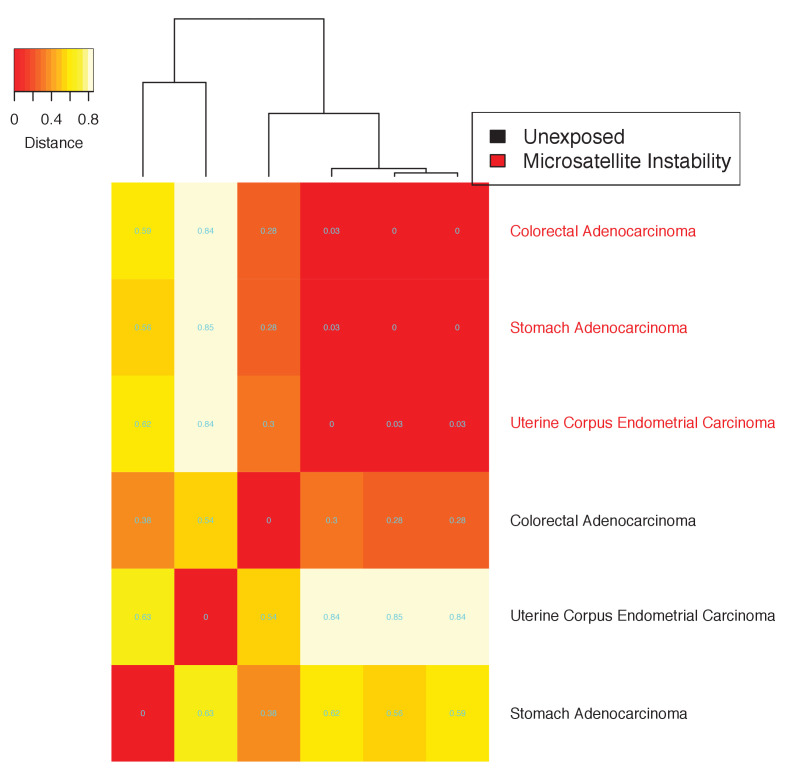
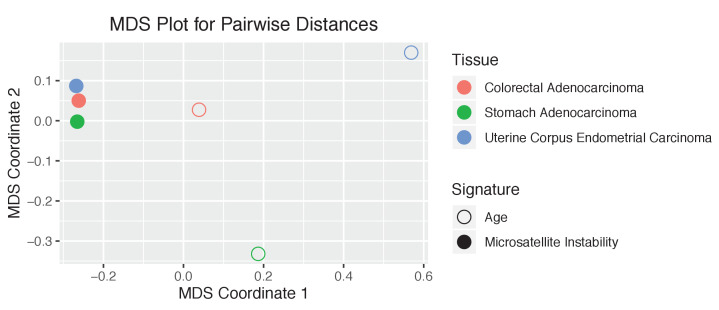
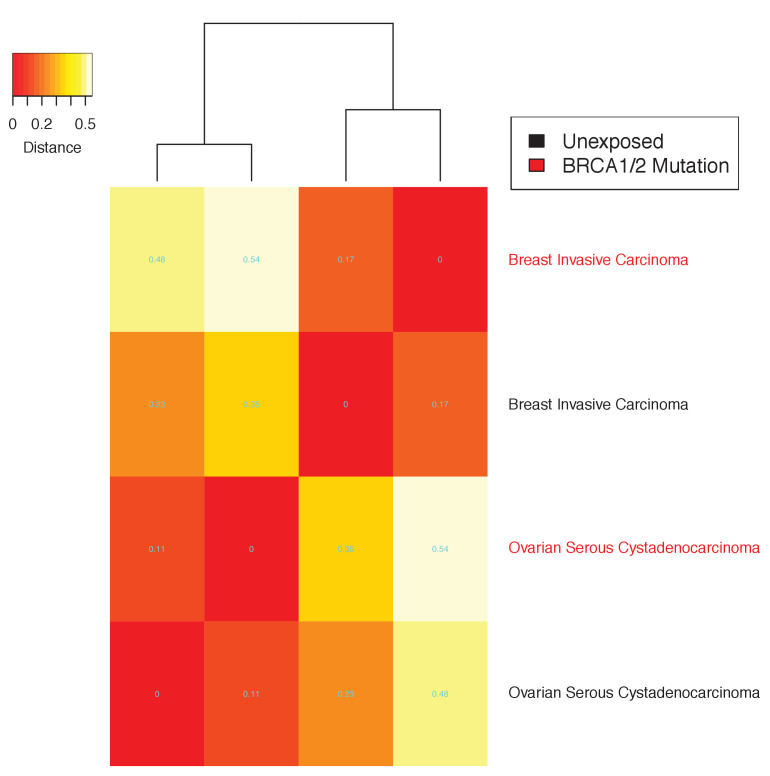
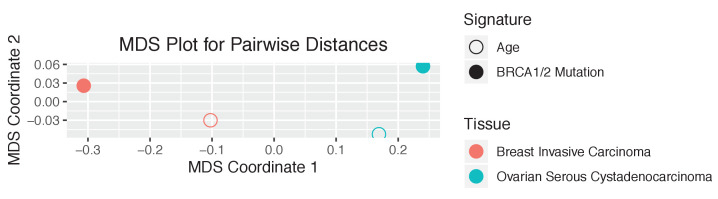
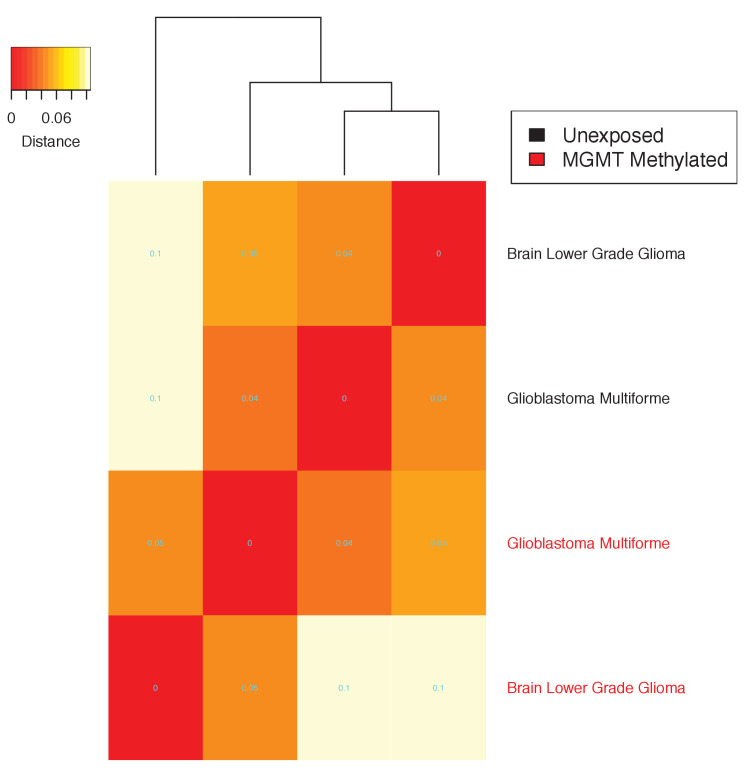
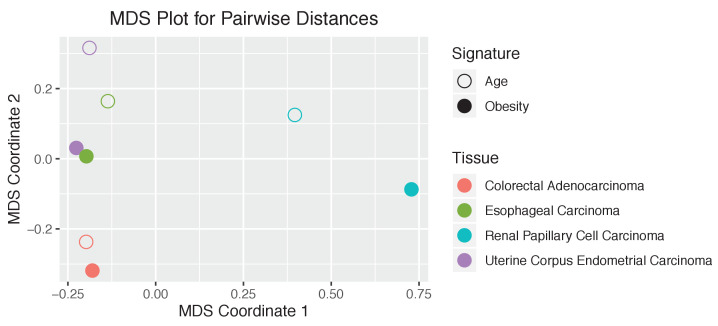
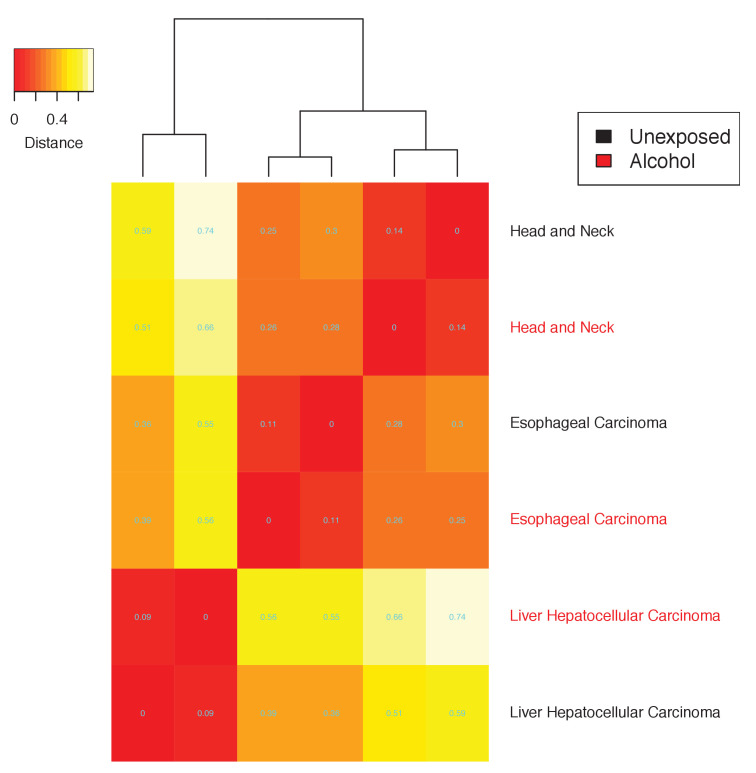
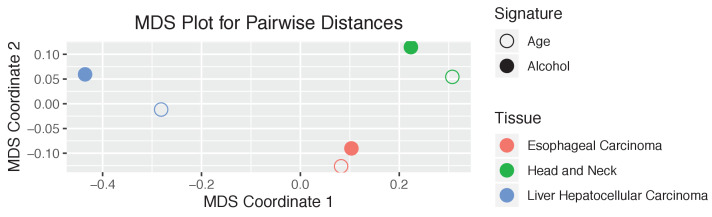
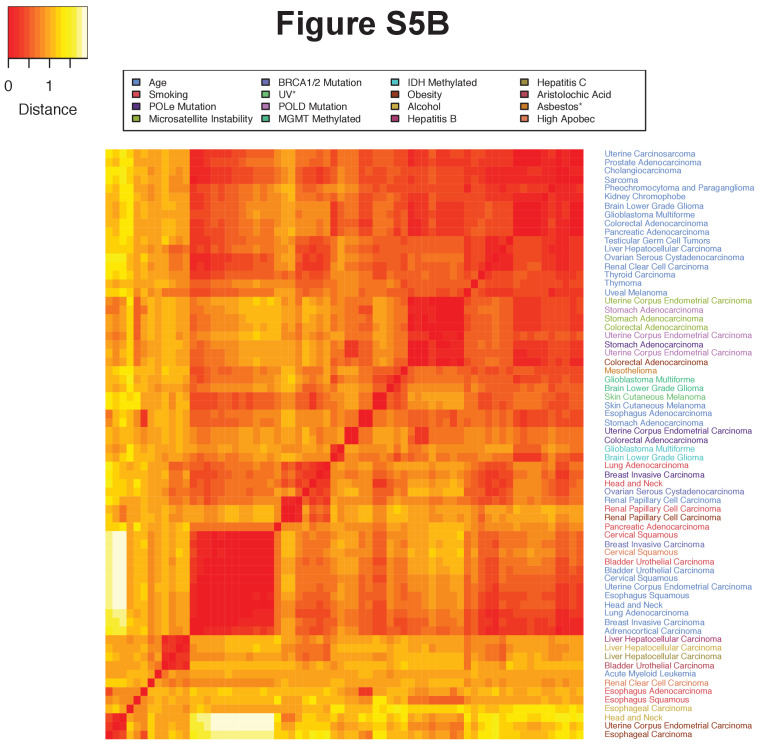
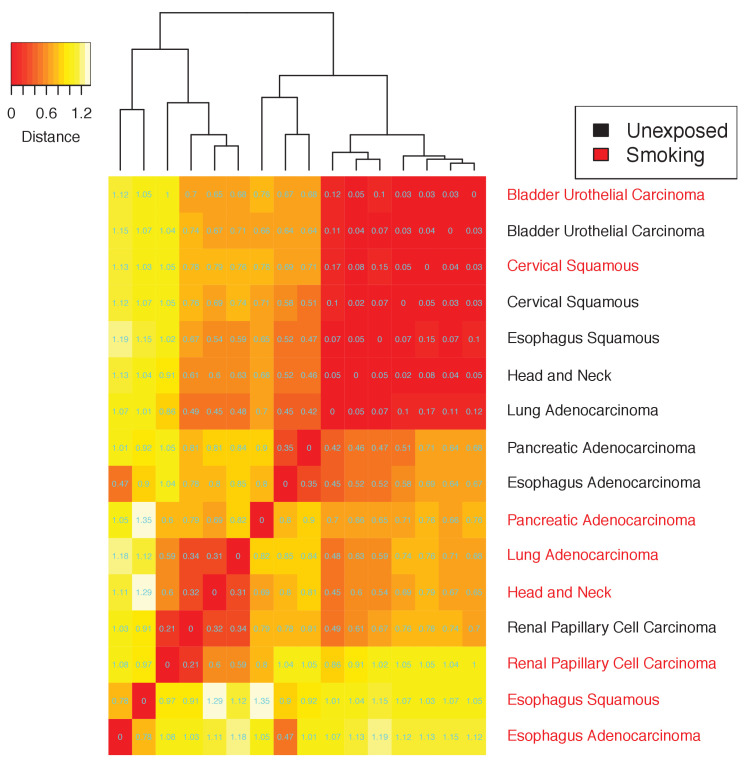
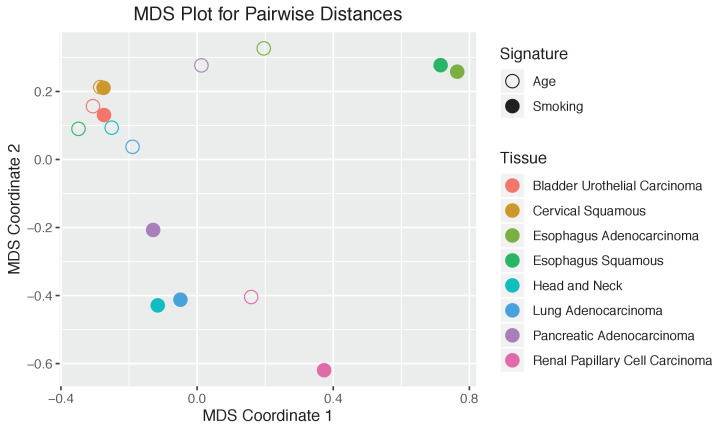
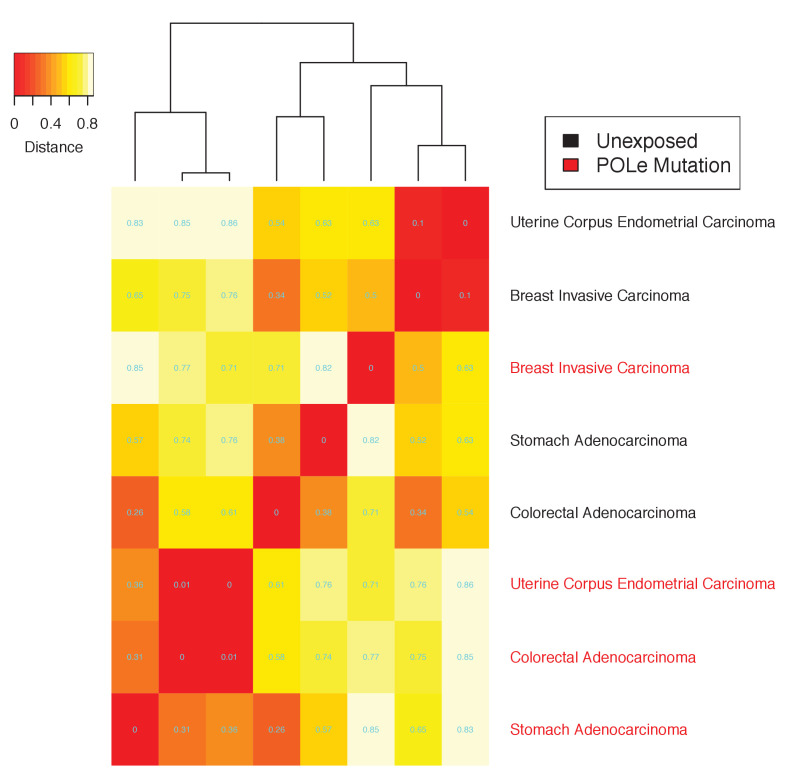
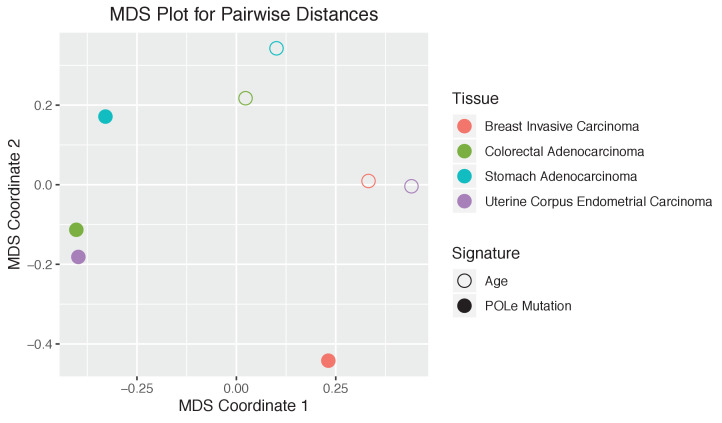
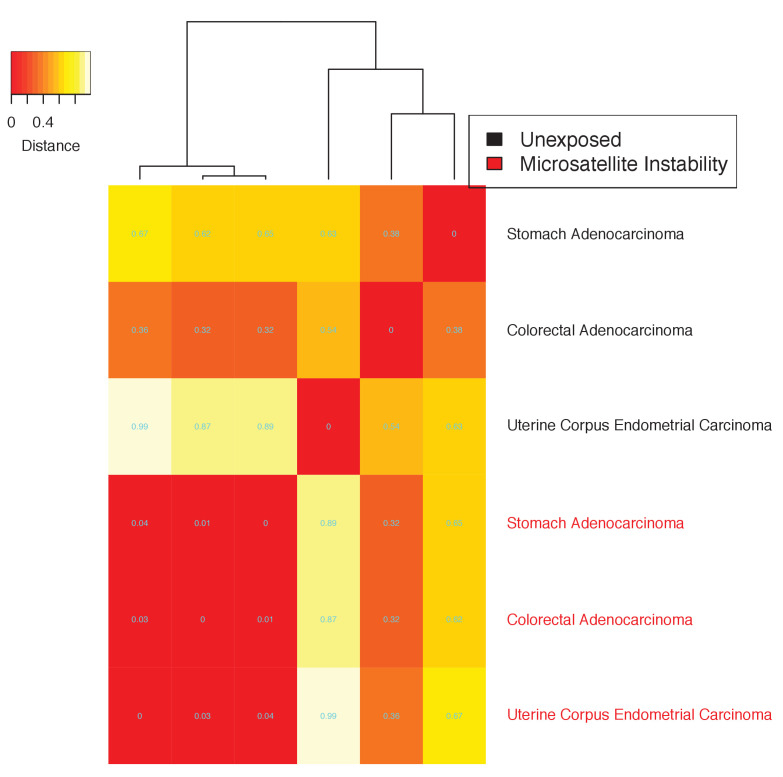
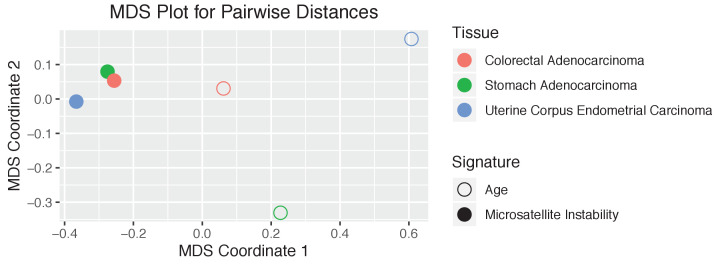
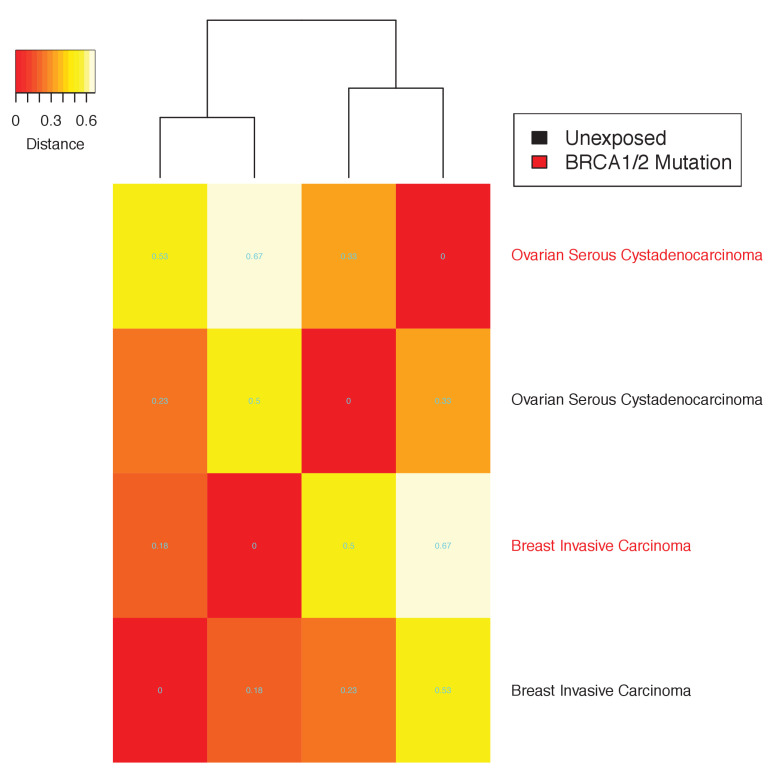
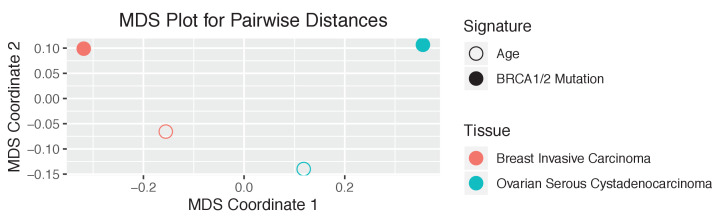
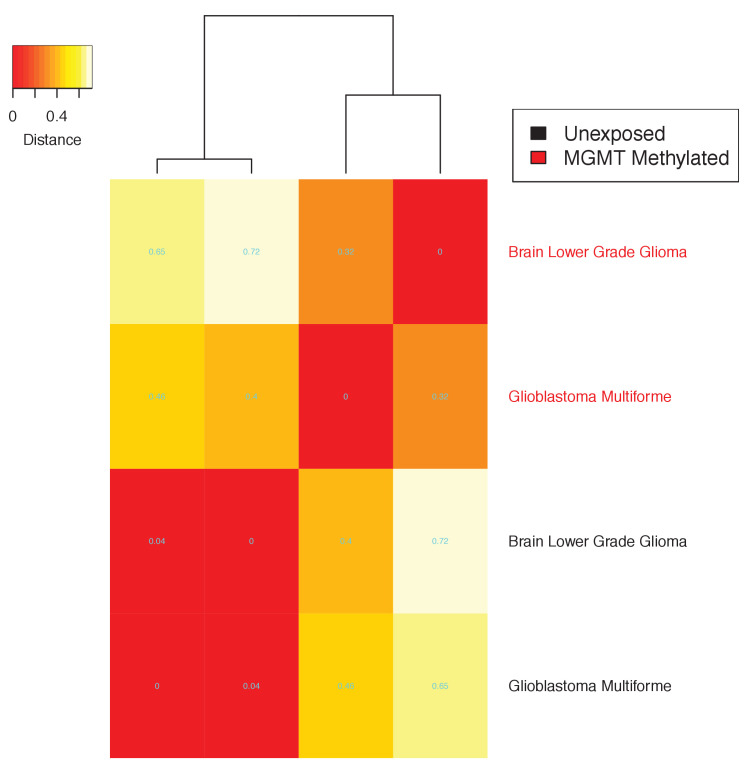
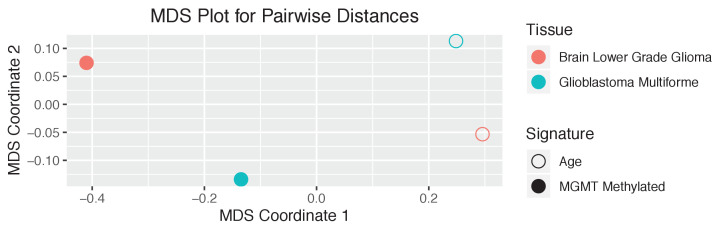
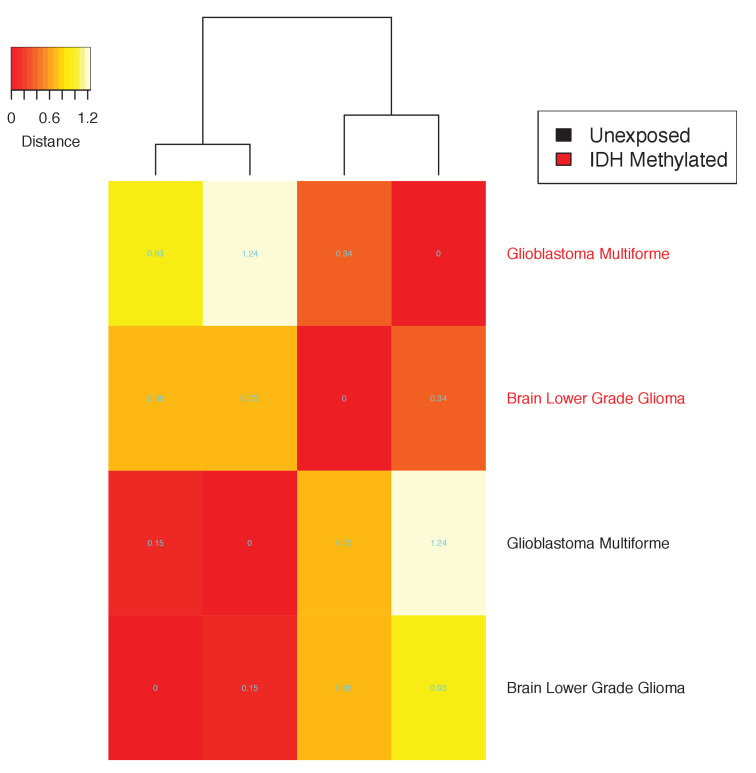
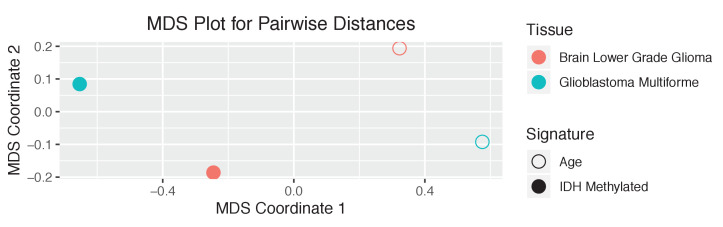
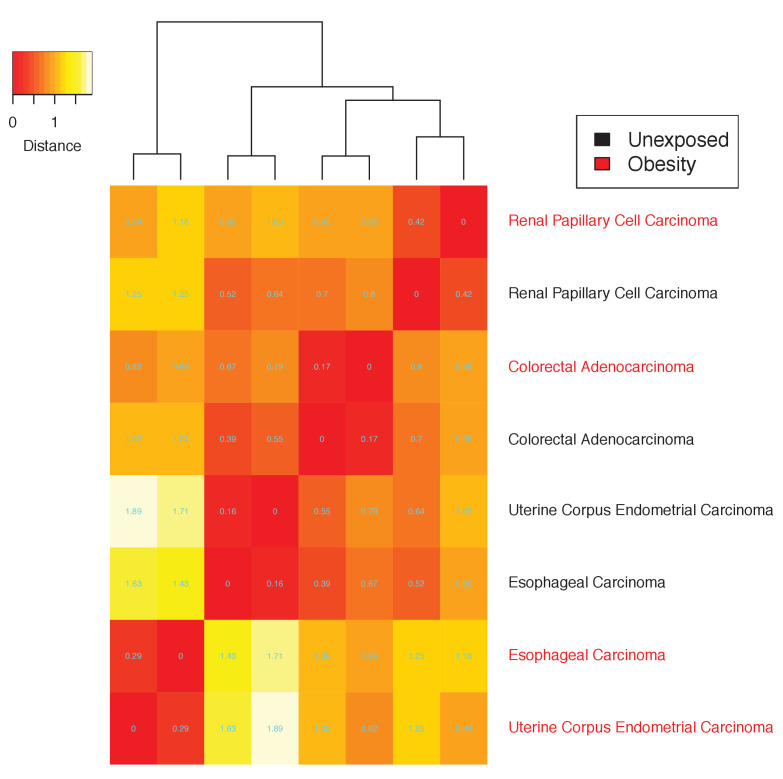
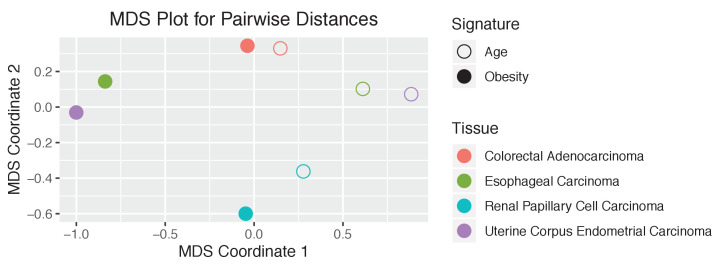
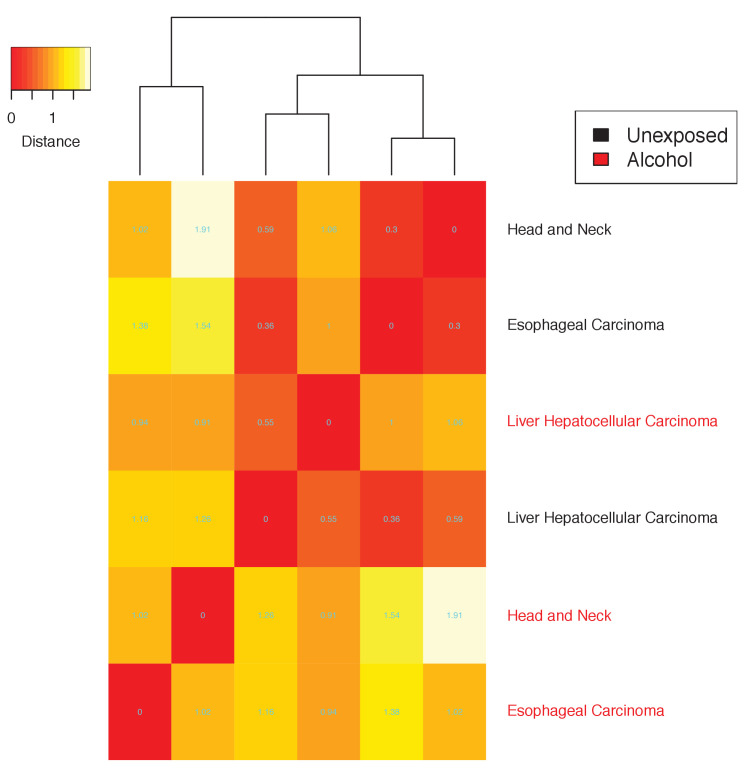
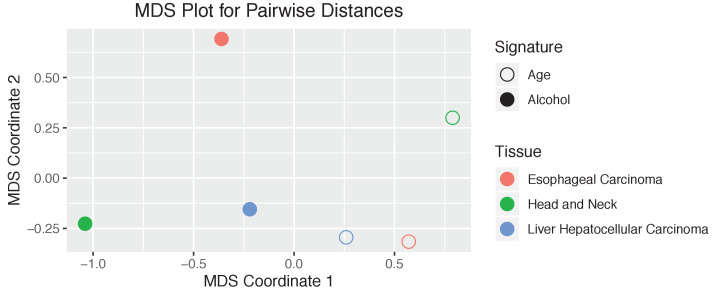
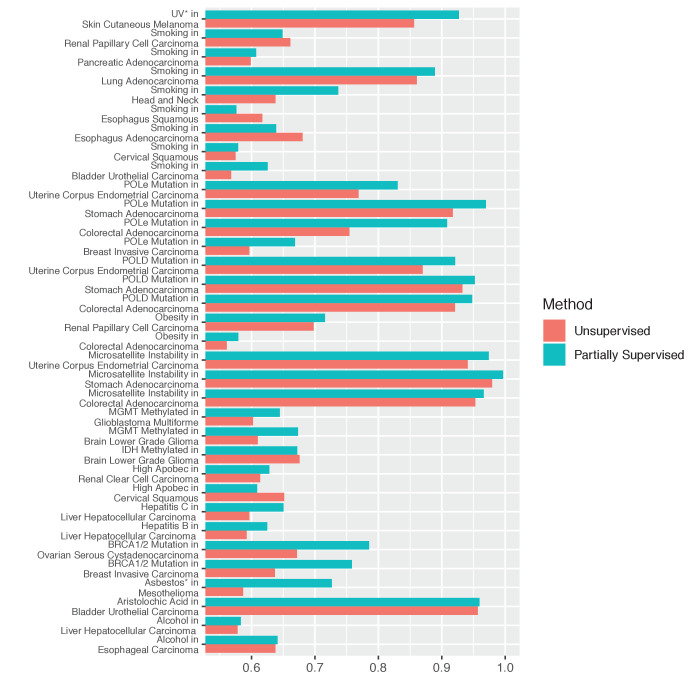
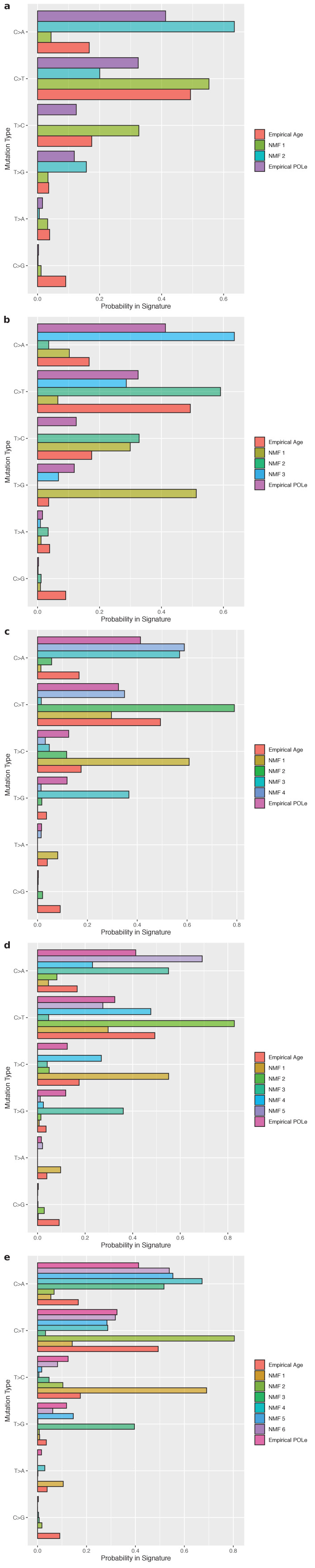
Figure 3. Comparisons of prediction accuracies (AUCs) of supervised, partially supervised, and unsupervised methodologies.
(a) Supervised age SuperSigs vs unsupervised Signature 1 over 30 tumor types; (b) SuperSigs vs unsupervised signatures for all annotated etiological factors other than age found in Alexandrov et al., 2013a, in tumor types for which the unsupervised signature was present (for the full list see Supplementary file 1). (c) Partially supervised vs unsupervised NMF signatures for all annotated etiological factors other than age (see Materials and methods). Each combination of tumor type and risk factor (e.g. lung adenocarcinoma and smoking) yields a signature and is represented by one point, which depicts the prediction accuracies of the unsupervised approach (x-axis coordinate value) versus the supervised (a–b) or partially supervised (c) one (y-axis coordinate value). Apparent AUCs are reported. The great majority (c) or essentially all (a–b) points lie above or on the line, indicating the greater accuracy of the supervised and partially supervised approaches.