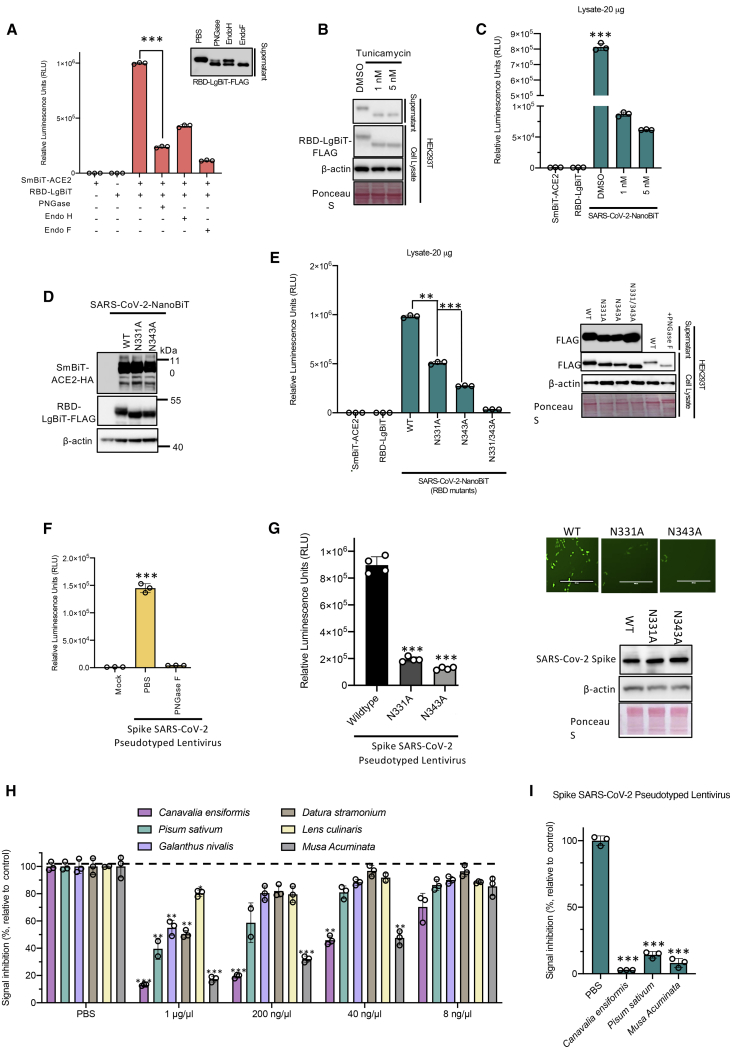
Figure 5.
N-linked glycosylation of SARS-CoV-2 S RBD is critical to its interaction with ACE2
(A) 293T cells transfected with either SmBiT-ACE2 or RBD-LgBiT were harvested. 25 μL of supernatants was pre-treated for 1 h at 37°C with Endo H, Endo F, PNGase F, or untreated, as indicated. Lysates were subsequently mixed and incubated for 15 min at room temperature, then assessed by luciferase assay using CTZ as a substrate (n = 3 biological replicates, mean ± SD; one-way ANOVA, ∗∗∗p < 0.005, Tukey’s correction for multiple comparisons). (B) 293T cells were transfected with RBD-LgBiT and 24 h after transfection were treated with tunicamycin for 16 h. Lysates and supernatants were prepared and analyzed by immunoblotting. (C) RBD-LgBiT cell lysates from (B) were incubated at room temperature with cell lysates from SmBiT-ACE2-transfected cells. After 15 min, the biosensor assay was performed. (D and E) Immunoblot analysis (D) and a bioreporter assay (E) were performed on cells co-transfected with the indicated RBD-glycosylation site mutants of the LgBiT-RBD constructs and SmBiT-ACE2 constructs. Immunoblot confirming equaling expression of Lg-RBD mutants is shown in right panel. (F) SARS-CoV-2 S pseudotyped lentivirus encoding ZsGreen and luciferase reporters was incubated with PBS ot or PNGase F for 1 h and then used to infect HEK293T-ACE2 cells. 48 h post-transduction, cells were evaluated for measuring luciferase activity. (G) SARS-CoV-2 S mutant pseudotyped lentiviruses infectivity assay as in (F). (H) Plant lectins were screened for the ability to disrupt CoV-NanoBiT. Cell lysates transfected with RBD-LgBiT were incubated for 1 h with different lectins from shown species. Luciferase assays were performed 5 min after SmBiT-ACE2 addition. (I) Plant lectins from Canavalia ensiformis (jack bean), Pisum sativum (pea), and Musa acuminata (banana) were evaluated for inhibitory effects against SARS-CoV-2 S pseudotyped lentivirus infection in HEK293T-ACE2 cells.