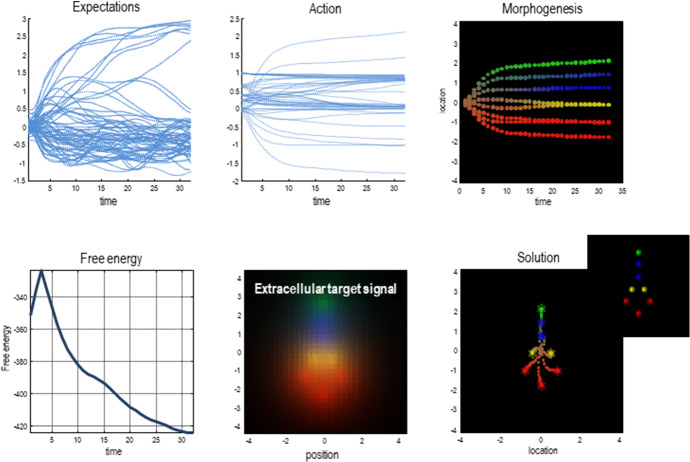
Fig. 6.
Multiscale self-organization and active inference. This figure depicts variational free energy being minimised across scales through active inference. It presents the results from a simulation of morphogenesis using the active inference framework (Friston et al. 2015). The simulation used a gradient descent on variational free energy to simulate a group of cells self-assembling into a larger pattern (i.e., target morphology). The simulation employed an ensemble of eight cells. Each cell was equipped with the same generative model, which is a metaphor for shared genetic information. This generative model generated a prediction of what each cell would sense and signal to other cells (chemotactically) for any given location in a target morphology. In other words, the model predicted what each cell would expect to sense and signal if it were in that location (lower middle panel—extracellular target signal). Each cell engaged in active inference, by actively moving around to infer its place in the target morphology relative to other cells. In doing so, each cell minimised its own variational free energy (and by proxy, its surprise or self-information). Remarkably, the fact that all cells shared the same generative model allowed their individual active inference to minimise the free energy of the ensemble, which exists at the scale above the individual cells. Each of the cells that make up the ensemble shares the same generative model. Crucially, the sensory evidence for the model with which each cell is equipped is generated by another cell. The arrangement that minimises the free energy of the ensemble is the target morphology. This means that each cell has to ‘find its place’; the configuration in which they all have found their place is the one where each cell minimises its own surprise about the signals it senses (because it knows its place), and in which the ensemble minimises the total free energy as well. The upper panels show the time courses of expectations about the place of each cell in the target morphology (upper left), as well as the associated active states that mediate cell migration and signal expression (upper middle). The resulting trajectories have been projected onto the first (vertical) direction and color-coded to show cell differentiation (upper right). The trajectories of each individual cell progressively, and collectively, minimize total free energy of the entire ensemble (lower left panel), which illustrates the minimization of free energy across scales. The lower right panel shows the configuration that results from active inference. Here, the trajectory is shown in small circles (for each time step). The insert corresponds to the target configuration. In short, all multiscale ensembles that are able to endure over time must destroy free energy gradients, which integrates system dynamics within and between scales.
Adapted from Friston et al. (2015). (Color figure online)