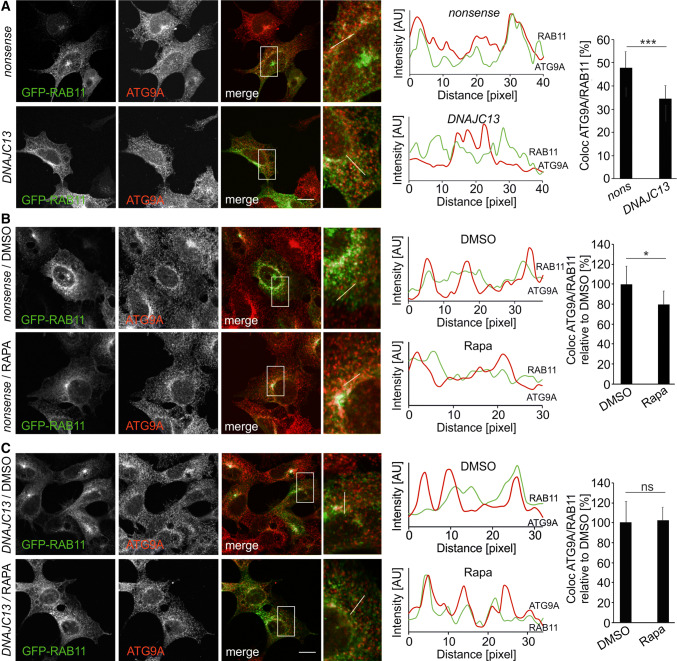
Fig. 3.
Knockdown of DNAJC13 alters localization of ATG9A at the recycling endosome. Confocal images of HEK293A cells transfected with nonsense or DNAJC13 siRNA and co-transfected with GFP-RAB11 under steady-state conditions (a) or rapamycin treatment (b, c). The box in each merged panel is enlarged and the fluorescent intensities along the white line are represented in the diagram, whereby ATG9A is shown in red throughout all panels. The overlap of ATG9A intensity peaks with GFP-RAB11 was quantified. The quantification method is described in detail in the Materials and Methods section. At least 13 regions of interest (ROIs) covering about 85 intensity peaks per cell were analyzed in 9–37 cells per condition (mean ± SD; t test: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001) (scale bar: 15 µM)