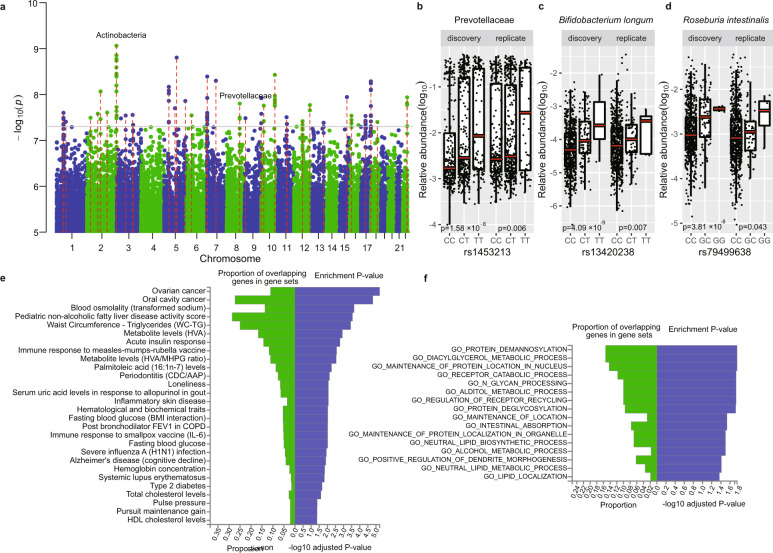
Fig. 2. M-GWAS results for the gut microbiome and functional analysis.
a Manhattan plot showing the associations between common variants and taxa, only suggestive associations with P < 10−5 were showed. The gray line represents a genome-wide significant P value (5 × 10−8). The red lines showed the 37 loci reaching the genome-wide significance. Two high heritable taxa, Actinobacteria linked to rs13420238 at LOC150935 and Prevotellaceae linked to rs1453123 at OXR1, were marked. b rs1453123 at OXR1 associated with Prevotellaceae both in discovery (P = 1.58 × 10−8) and replication (P = 0.006) cohort. c rs13420238 at LOC150935 associated with Bifidobacterium longum (Actinobacteria) both in discovery (P = 4.09 × 10−9) and replication (P = 0.007) cohort. d rs79499638 near PLXDC2 associated with Roseburia intestinalis both in discovery (P = 3.81 × 10−8) and replication (P = 0.043) cohort. e Enriched traits or diseases identified in GWAS catalog using 109 mapped genes from 37 significant loci. f Enriched KEGG or GO pathways identified for 109 mapped genes from 37 significant loci.