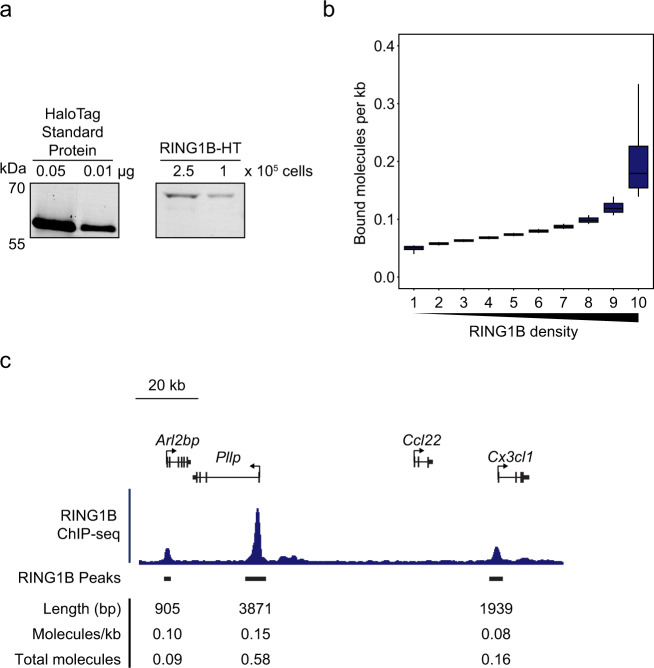
Fig. 3. RING1B has low target site occupancy.
a An example of an in-gel fluorescence image of TMR-labelled HaloTag Standard Protein loaded in known quantities (left panel) and RING1B-HT-TMR from whole-cell lysates of a known number of cells (right panel). Fluorescence intensity from n = 3 biological replicates was used to quantify the number of molecules present81. Source data are provided as a Source Data file. b A box plot illustrating the number of RING1B molecules that are estimated to be bound per kilobase of DNA at RING1B peaks from ChIP-seq in ESCs79. Read distributions aggregated from n = 3 biological replicates of RING1B ChIP-seq were used to segregate bound RING1B molecules into density deciles over RING1B peaks (18,643). Boxes represent the interquartile range (IQR), the middle line corresponds to the median, and whiskers extend to the largest and smallest values no more than 1.5 x IQR from the box. Values outside of this range are not plotted, but are included in all analyses. Source data are provided as a Source Data file. c A genomic snapshot of RING1B ChIP-seq illustrating the varying distribution of RING1B binding between individual peaks. The values shown below indicate the length of the RING1B peak, the estimated density of RING1B molecules bound at each peak, and the estimated number of molecules bound per allele to each peak assuming a 2n genome.