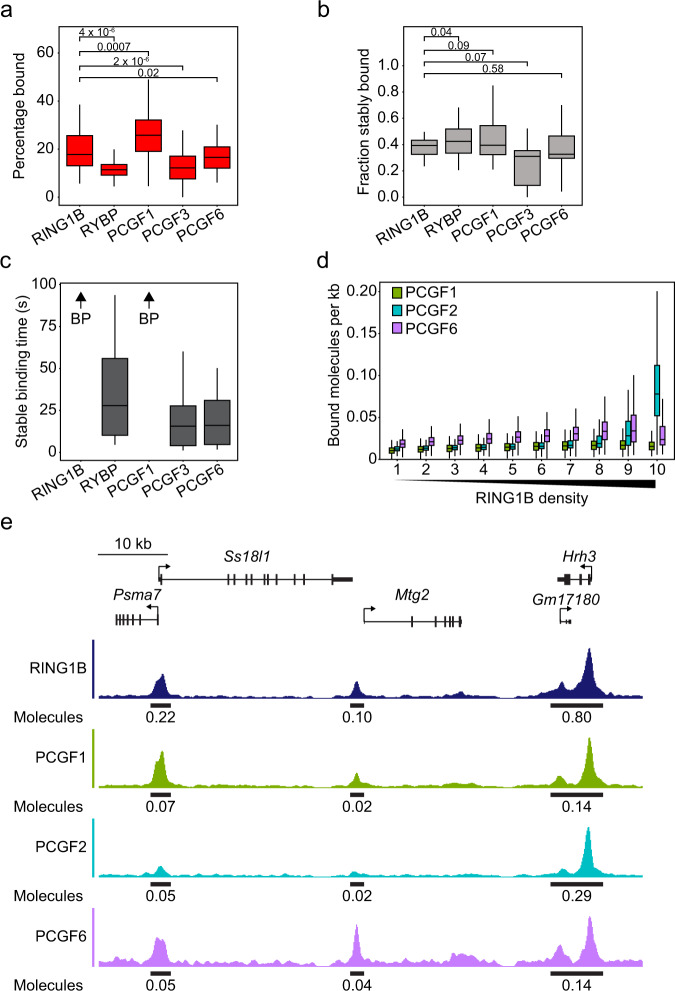
Fig. 7. Variant PRC1 subunits exhibit distinct dynamics and broad binding to PRC1 targets.
a A box plot indicating the percentage of RING1B, RYBP, PCGF1, PCGF3 and PCGF6 molecules in the bound state determined by SPT. Box plots represent measurements from n > 50 movies from 3 experiments each. Indicated p-values were calculated using a two-tailed Student’s t test. Source data are provided as a Source Data file. b A box plot indicating the fraction of RING1B, RYBP, PCGF1, PCGF3 and PCGF6 molecules that exhibit stable binding. Box plots represent measurements from n > 15 movies from at least 2 experiments each. Indicated p-values were calculated using a two-tailed Student’s t test. Source data are provided as a Source Data file. c A box plot indicating the stable binding times for RING1B, RYBP, PCGF1, PCGF3 and PCGF6 after photobleaching correction in 2 Hz tracking experiments. BP (beyond photobleaching) and an arrow indicates proteins for which stable binding exceeded the photobleaching time. Box plots represent measurements from n > 15 movies from at least 2 experiments each. Source data are provided as a Source Data file. d A box plot illustrating the number of PCGF1 (green), PCGF2 (turquoise), PCGF6 (purple) molecules that are estimated to be bound per kilobase of DNA at RING1B peaks from ChIP-seq in ESCs79. Read distributions aggregated from n = 3 biological replicates of corresponding ChIP-seq were used to segregate bound molecules into density deciles over RING1B peaks. Source data are provided as a Source Data file. e A genomic snapshot of RING1B, PCGF1, PCGF2 and PCGF6 ChIP-seq illustrating the differing distributions of PRC1 complexes across RING1B peaks. Values shown below the ChIP-seq signal indicate the estimated number of RING1B, PCGF1, PCGF2 and PCGF6 molecules bound at a single allele for the indicated peak. In a–d, boxes represent the interquartile range (IQR), the middle line corresponds to the median, and whiskers extend to the largest and smallest values no more than 1.5 x IQR from the box. Values outside of this range are not plotted, but are included in all analyses.