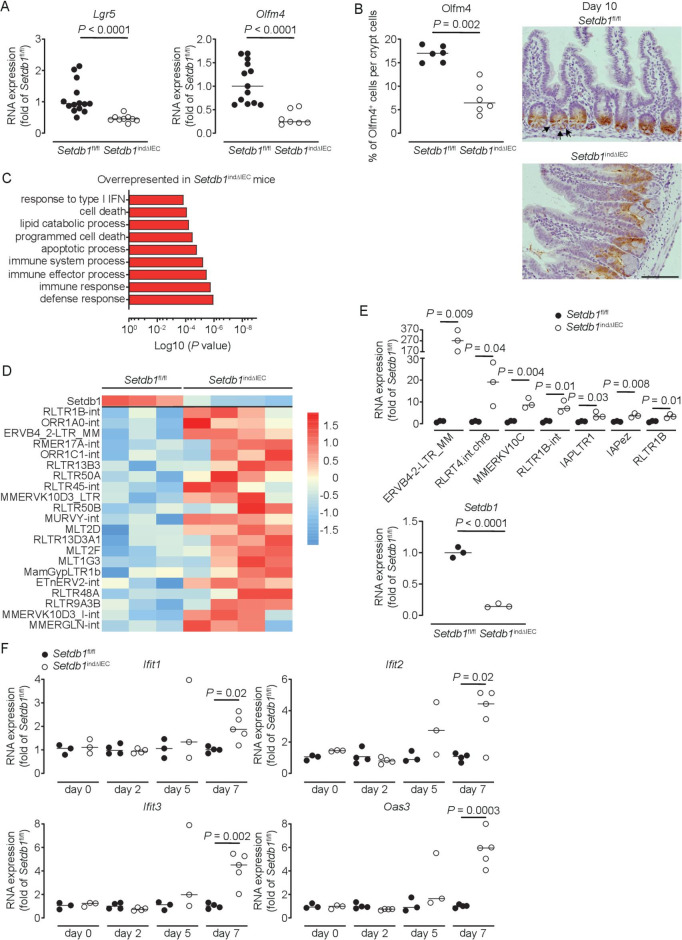
Figure 4.
Setdb1 deletion leads to de-repression of endogenous retroviruses and induction of a type I interferon response. (A) RNA expression of Lgr5 and Olfm4 as determined by quantitative PCR (qPCR) in the ileum of the indicated mouse strains 9 days after the first tamoxifen injection. (B) Quantification and representative immunohistochemistry staining of OLFM4 in the ileum of the indicated mouse strains 10 days after the first tamoxifen injection. Arrows show intestinal stem cells (ISCs) in Villin-CreERT2-negative Setdb1 fl/fl mice. (C) Gene ontology (GO) terms of the top 100 upregulated genes in RNA sequencing of small intestinal crypts of Setdb1 indΔIEC mice 2 days after the first tamoxifen injection. (D) Heatmap of the expression of Setdb1 (top) and endogenous retroviruses (ERVs) (below) as obtained from RNA sequencing of small intestinal crypts of Setdb1 indΔIEC mice 2 days after the first tamoxifen injection. (E) Expression of the indicated ERVs (top) and Setdb1 (below) in the ileum of the indicated mouse strains 5 days after the first tamoxifen injection as determined by qPCR. (F) RNA expression of the indicated genes as determined by qPCR in the small intestine of mice at the indicated time points after the first tamoxifen injection. Representative results of two-three independent experiments are shown (A–B, E–F). RNA sequencing was performed once (C–D). In (A–B, E–F), dots represent individual mice and the bar indicates the median. Size bars indicate 100 µm. The Mann-Whitney U test (A–B, E–F) was applied. SETDB1, SET Domain Bifurcated Histone Lysine Methyltransferase 1, also known as ESET.