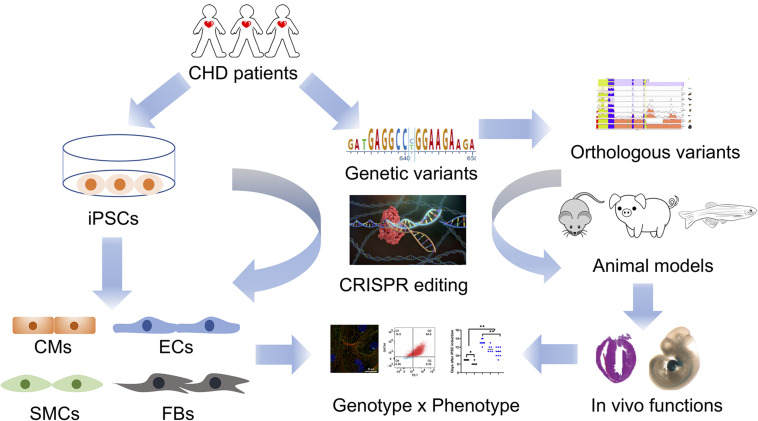
FIGURE 1.
An integrated patient-specific iPSC model for studying genetics of CHD. Whole genome sequencing of CHD patients identifies prospective genetic variants that are retained in patient-specific iPSCs. Cardiovascular defects in CHD are recapitulated in patient iPSC-derived cardiac cells, such as cardiomyocytes (CMs), endocardial/endocardial cells (ECs), vascular smooth muscle cells (SMCs), and fibroblasts (FBs). Genetic variants associated with a CHD phenotype are corrected in diseased iPSCs and introduced to healthy iPSCs using CRISPR/Cas9 genome editing tools. These genome-edited iPSCs are further investigated to validate the cause-effect relationship between a given genetic variant and a CHD phenotype. In parallel, orthologous variants are genetically introduced into animal models (rodents, pigs, zebrafish, etc.) in order to investigate the in vivo effects of a given human variant on heart development. Together, an integrated model including both genome-edited human iPSCs and transgenic animals will yield a more comprehensive illustration of the genetic basis of CHD in the new era of genomic medicine.