Abstract
The carbohydrate-binding protein LecA (PA-IL) from Pseudomonas aeruginosa plays an important role in the formation of biofilms in chronic infections. Development of inhibitors to disrupt LecA-mediated biofilms is desired but it is limited to carbohydrate-based ligands. Moreover, discovery of drug-like ligands for LecA is challenging because of its weak affinities. Therefore, we established a protein-observed 19F (PrOF) nuclear magnetic resonance (NMR) to probe ligand binding to LecA. LecA was labeled with 5-fluoroindole to incorporate 5-fluorotryptophanes and the resonances were assigned by site-directed mutagenesis. This incorporation did not disrupt LecA preference for natural ligands, Ca2+ and d-galactose. Following NMR perturbation of W42, which is located in the carbohydrate-binding region of LecA, allowed to monitor binding of low-affinity ligands such as N-acetyl d-galactosamine (d-GalNAc, Kd = 780 ± 97 μM). Moreover, PrOF NMR titration with glycomimetic of LecA p-nitrophenyl β-d-galactoside (pNPGal, Kd = 54 ± 6 μM) demonstrated a 6-fold improved binding of d-Gal proving this approach to be valuable for ligand design in future drug discovery campaigns that aim to generate inhibitors of LecA.
Keywords: drug discovery, LecA, lectin, NMR
Introduction
Many opportunistic pathogens, such as the Gram-negative bacterium Pseudomonas aeruginosa, use glycan-binding proteins (lectins) to infect the host and to establish a protective antibiotic-resistant biofilm environment in the lungs of immunocompromised patients (Singh et al. 2000). LecA plays a key role in this process and has become a promising target to prevent biofilm formation and consequently disease progression (Diggle et al. 2006; Wagner et al. 2016).
LecA forms a protein homotetramer (Fig. 1A) and requires a calcium (II) ion (Ca2+) to coordinate binding to its natural monosaccharide ligand, d-galactose (d-Gal) (Cioci et al. 2003). Notably, each monomer has four tryptophan residues (Fig. 1B), where W42 and W33 reside in close proximity to the d-Gal-binding region and near the hinge connecting the two domains, respectively (Fig. 1C). In the physiological context of a biofilm, the low micromolar affinity of LecA for d-Gal (Kd = 88 μM, (Kadam et al. 2011)) is compensated with high avidity of ligands that can crosslink the neighboring carbohydrate-binding sites, such as GalAxG3 (Kd = 2.5 nM, (Bergmann et al. 2016; Cecioni et al. 2015)); however, drug-like molecules do not benefit from such multivalency. Successful approaches to find low-molecular-weight inhibitors have capitalized on the β-linked galactosides with aromatic aglycon (Garber et al. 1992), such as p-nitrophenyl β-d-galactoside (pNPGal) or GalAG0 with Kd = 14.1 μM and Kd = 4.2 μM, respectively (Kadam et al. 2011; Kadam et al. 2013; Rodrigue et al. 2013). This improvement in binding affinity is due to the additional CH–π interaction between the aromatic ring of the binding ligand and H50 at the binding pocket. Cumulatively, development of inhibitors for LecA consists of β-linked carbohydrate-based ligands (glycomimetics, (Wagner et al. 2017)) and, therefore, having drug-like ligands for LecA is crucial in future research, but discovery of such weak binders is challenging. Therefore, new methods to detect binding of weak ligands to LecA are required.
Fig. 1.
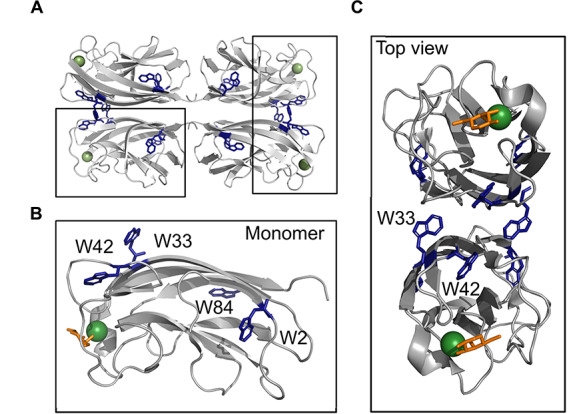
Structure of LecA. Cartoon representation of tetramer LecA (PDB: 4CP9). Shown is an expansion of a LecA monomer with D-Gal (shown as sticks), Ca2+ ion (shown as sphere) and positions of four tryptophanes (W2, W33, W42, W84). W2 and W84 are in the protein core. Top view shows W42 and W33 being located near the carbohydrate-binding site of LecA.
Biophysical methods are suitable to identify low-molecular-weight and low-affinity ligands (Renaud et al. 2016). 19F nuclear magnetic resonance (NMR) has proven to be valuable in the study of protein–ligand interactions for several reasons. The isotope 19F 1) has the spin 1/2 nucleus and a natural abundance of 100%, 2) is very stable and 3) nearly absent in biological systems delivering a background-free NMR spectrum (Luck and Falke 1991). In the case of 19F-labeled proteins used in protein-observed 19F (PrOF) NMR, the size of protein is not a limitation and the protein side chains are detected as broad resonances at low (25 μM) to mid-micromolar (200 μM) protein concentrations (Kitevski-LeBlanc and Prosser 2012; Liu et al. 2012). This method has benefited from the commercial availability of many fluorinated aromatic amino acids, such as 5-fluorotryptophan (5FW), 3-fluorotyrosine and 4-fluorophenylalanine. Unfortunately, these fluorine-labeled amino acids are expensive. In contrast, fluorine-labeled precursors of amino acids, such as 5-fluoroindole (5FI), can be employed to incorporate fluorine-labeled amino acids in proteins, resulting in reduced costs (Gee et al. 2016). Moreover, incorporation of fluorinated amino acids does not lead to major structural and functional perturbations (Arntson and Pomerantz 2016; Kitevski-LeBlanc and Prosser 2012; Sharaf and Gronenborn 2015). In this context, 5FW has been shown to have only a minor impact on protein structure and dynamics in bacterial lectin from Ralstonia solanacearum lectin (RSL) (Tobola et al. 2018).
Here, we explored PrOF NMR using LecA labeled with 5FW (5FW LecA) to detect binding of ligands with moderate as well as low affinities. To assign 5FW resonances, we produced its wild-type (WT) and four tryptophan-to-phenylalanine mutants (W2F, W33F, W42F and W84F). In the binding studies, we determined the dissociation constants of 5FW LecA with its natural ligands Ca2+, d-Gal and d-GalNAc. We compared the affinity data of LecA and 5FW LecA with other orthogonal biophysical methods, such as isothermal titration calorimetry (ITC) or competitive binding by fluorescence-polarization (FP) detection. Finally, we verified the suitability of 5FW LecA PrOF NMR for a ligand design using glycomimetics pNPGal and phenyl-β-d-galactopyranoside (Ph-β-d-Gal, (Imberty et al. 2004)).
Results and discussion
Protein expression and characterization
For the stable incorporation of 5FW in LecA we followed the workflow shown in Fig. 2A. Escherichia coli BL21 (DE3) cells were grown in presence of 5FI and the protein was characterized for fluorine incorporation via mass spectrometry (Fig. 2B and C). In the mass spectrum 5FW LecA had a dominant mass of 12831.34 Da corresponding to full incorporation of four tryptophan residues being replaced with 5FW. Protein yields as high as 45–50 mg L−1 using non-auxotrophic E. coli BL21 (DE3) cells were achieved. This compares very well to protein expression yields under non-labeling conditions (30–35 mg L−1).
Fig. 2.
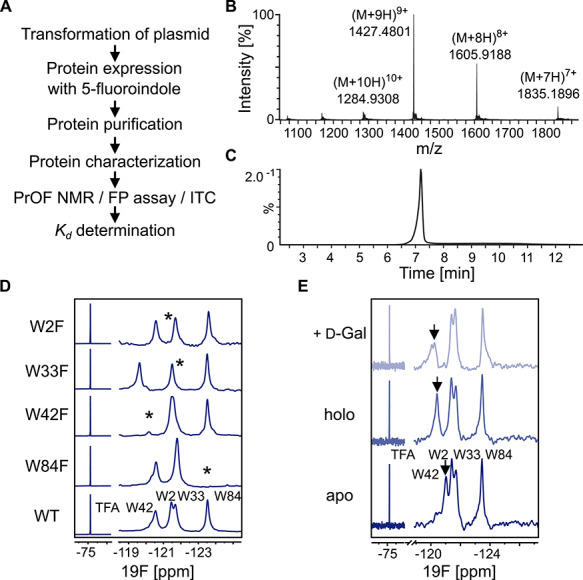
PrOF NMR of 5FW LecA. (A) General workflow for PrOF NMR with 5FW LecA. (B) Chromatogram of the LC–ESI–MS analysis of 5FW LecA. (C) ESI-MS+ spectrum of the main peak at 7.3 min [M + H]+Ca = 12826.23 Da [M + H]+found = 12831.34 Da corresponds to 5FW LecA. (D) PrOF NMR assignment of 5FW LecA WT and the mutants W84F, W42F, W33F and W2F. The tryptophanes being mutated are indicated with asterisk. All spectra were normalized and referenced to TFA. (E) PrOF NMR of 5FW LecA WT in Ca2+-free (apo, bottom) and -bound (holo, central) forms. The W42 resonance (black arrow) shifted in presence of Ca2+ and 0.5 mM d-Gal binding verifying that protein is active.
Establishing 5FW LecA in PrOF NMR
To optimize PrOF NMR for 5FW LecA, we varied temperature (Supplementary Fig. S1), protein concentration (Supplementary Fig. S2) and buffer (Supplementary Fig. S3). Fluorine resonances of 5FW LecA resulted in a well-resolved PrOF NMR spectrum using 100–200 μM 5FW LecA at 310 K on a 700 MHz NMR machine equipped with a cryogenic probe (Supplementary Table SI).
Next, we assigned the fluorine resonances in PrOF NMR to four tryptophanes of LecA using site-directed mutagenesis (Fig. 2D). The disappearance of fluorine resonances in LecA mutants being replaced with phenylalanine gave indication for the assignment. As result, two peaks at −120.47 and −123.24 p.p.m. could be reliably identified as W42 and W84, respectively. Two resonances at −121.43 and −121.72 p.p.m. overlapped slightly indicating that both reside in a similar chemical environment in LecA, but could be assigned as W2 and W33, respectively.
Incorporation of 5FW into LecA does not abrogate protein activity
Because in some cases the affinity of proteins to the carbohydrates might be changed due to 5FW incorporation, as it has been shown for a stronger affinity of 5FW to β-d-Gal (Hudson et al. 2015), we confirmed that incorporation of 5FW in LecA does not introduce structural or functional perturbations that could alter protein activity. For this, we measured 5FW LecA in PrOF NMR in its Ca2+-free (apo), -bound (holo) and d-Gal-bound forms (Fig. 2E). As the preference of LecA for both Me-α-d-Gal and Me-β-d-Gal has been reported (Kd = 50 μM and 55.7 μM, respectively; Rodrigue et al. 2013), suggesting that the anomeric configuration of d-Gal is not critical, we used d-Gal containing both anomeric configurations to verify the protein activity. As a result, 19F resonance of W42 showed a chemical shift perturbation (CSP) upon binding to both Ca2+ and d-Gal indicating that 5FW LecA remained in its active form.
Next, we measured the affinities of 5FW LecA to its natural ligands. For this, we titrated Ca2+ (Supplementary Fig. S4) and d-Gal (Supplementary Fig. S5) to 5FW LecA, which resulted in binding affinities Kd of 47 8 μM and 360
8 μM and 360 47 μM, respectively. Despite the difference to previously reported affinity for d-Gal (Kadam et al. 2011), the 2- or 3-fold deviation in binding affinities determined in PrOF NMR has been considered acceptable in PrOF NMR (Gee et al. 2016; Tobola et al. 2018). In our experience, we have considered a 4-fold change acceptable to continue with affinity assessment.
47 μM, respectively. Despite the difference to previously reported affinity for d-Gal (Kadam et al. 2011), the 2- or 3-fold deviation in binding affinities determined in PrOF NMR has been considered acceptable in PrOF NMR (Gee et al. 2016; Tobola et al. 2018). In our experience, we have considered a 4-fold change acceptable to continue with affinity assessment.
Next, we confirmed the affinities for Ca2+ and d-Gal with both LecA and 5FW LecA in ITC (Supplementary Fig. S6) and a competitive binding fluorescence polarization (FP) assay, respectively (Supplementary Fig. S7; Joachim et al. 2016). As a result, binding experiments of 5FW LecA with Ca2+ and d-Gal confirmed the affinities to be in similar range with LecA (Supplementary Table SIV), concluding that 5FW LecA preserved its activity and preference to its natural ligands similarly to LecA.
PrOF NMR to probe weak LecA–ligand interactions
To establish a method for the discovery of drug-like molecules for LecA, our aim was to probe 5FW LecA in PrOF NMR for binding of a known weak ligand. For this, we chose d-GalNAc (Fig. 3A; Chemani et al. 2009). We observed that d-GalNAc perturbed W42 resonance located in the carbohydrate-binding site of 5FW LecA. The changes in W42 peak intensity (Fig. 3B) upon addition of d-GalNAc were followed to derive the Kd value of 780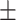 97 μM (Fig. 3C).
97 μM (Fig. 3C).
Fig. 3.
PrOF NMR to probe weak 5FW LecA–ligand interactions. (A) Structure of N-acetyl d-galactosamine (d-GalNAc). (B) The PrOF NMR spectra of holo 5FW LecA (bottom) and titration of d-GalNAc (upper). d-GalNAc binding affects W42 the strongest and thus, we followed the changes in peak intensity of free W42 to derive Kd values for d-GalNAc binding. (C) Binding isotherm for d-GalNAc generated by plotting the normalized change in peak intensity of 5FW free W42 resonance as a function of ligand concentration. Data of three independent titrations were fitted to one-site-binding model to obtain Kd of 780 ± 97 μM.
Similarly as before, we compared the affinities of 5FW LecA for d-GalNAc in a FP-based assay and the IC50 was 3-fold higher compared with the Kd obtained from PrOF NMR confirming that d-GalNAc is much weaker ligand compared with Ca2+ or d-Gal. Moreover, our affinity data in the FP assay for ligands, in particular d-Gal, were in a close range 1230 ± 200 μM and 1991 μM for both unlabeled LecA and 5FW LecA, respectively (Supplementary Table SIV). Cumulatively, this result suggests that the affinities for d-GalNAc derived from the FP assay for LecA and 5FW LecA diverged from PrOF NMR because of higher sensitivity of 19F NMR to spot weak binders and thus, thereby shows the advantages of PrOF NMR in discovery of weak interactions.
5FW LecA PrOF NMR is sensitive to probe glycomimetics
PrOF NMR with 5FW can be useful for discovery and design of ligands for LecA. For this, we performed PrOF NMR titrations of two glycomimetics: phenyl-Ph-β-d-Gal (Supplementary Fig S6) and pNPGal (Fig. 4) to 5FW LecA resulting in Kd of 166 ± 42 μM and 54 ± 6 μM, respectively. Moreover, p-nitrophenyl group improved binding affinity of d-Gal 6-fold, which is in agreement with previous reports (Rodrigue et al. 2013). This shows that 5FW in LecA can serve as sensitive probes to follow the affinity gain to design glycomimetics using structure-activity relationship approach (Divakaran et al. 2019).
Fig. 4.
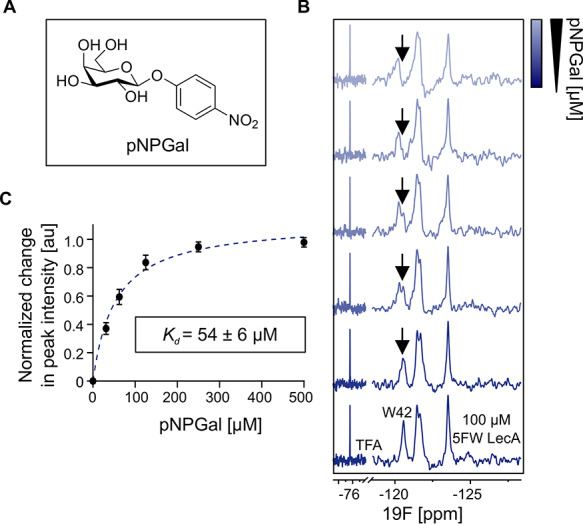
PrOF NMR titration of a carbohydrate-based glycomimetic pNPGal to holo 5FW LecA. (A) The structure of pNPGal. (B) The PrOF NMR spectra of holo 5FW LecA (bottom) and titration of pNPGal (upper). The peak intensity of W42 resonance (arrow) decreased upon pNPGal addition. The change in signal intensity of free W42 peak can be followed to determine Kd. (C) Binding isotherm for pNPGal generated by plotting the normalized change in peak intensity of 5FW free W42 resonance as a function of ligand concentration. Data of three independent titrations were fitted to one-site-binding model to obtain Kd of 54 ± 6 μM.
Conclusions
We have shown that 5-fluoroindole can be used as a precursor of 5FW to label LecA for PrOF NMR studies. In our binding studies with Ca2+, d-Gal and d-GalNAc, PrOF NMR has proven to detect and determine the affinity of moderate as well as weak ligands. In contrast to ligand-observed NMR techniques (e.g. STD NMR; Mayer and Meyer 2001) providing information on the epitope of ligand binding, PrOF NMR provides information on the ligand-binding site in the protein.
Further studies using ITC and FP assays have demonstrated that 5FW LecA preserved its activity and the ligand preference similarly to LecA. Notably, PrOF NMR has proven more sensitive for identification of weak ligands like d-GalNAc due to chance to observe the formation of a protein–ligand complex in NMR at earlier time point compared with the FP assay. Accordingly, these results represent the first studies demonstrating the potential of 5FW LecA PrOF NMR to assess binding of weak ligands. As tryptophan is by far the most frequently found amino acid in carbohydrate binding sites of various lectins (Taroni et al. 2000), this method could prove to be a valuable tool to assess binding of fragment- and drug-like molecules targeting the carbohydrate binding site of various lectins. Together, this approach will support the future drug-discovery campaigns that aim to develop drug-like inhibitors for lectins such as LecA.
Materials and methods
Fluorinated protein expression and purification
Recombinant 5FW LecA (WT and mutants) was expressed and purified as follows: E. coli BL21 (DE3) cells were transformed with pET25pa1l plasmid and grown in LB medium (100 μg mL−1 ampicillin) at 37°C with agitation (120 rpm) until OD600 reached 0.6. 1 L of culture was harvested by centrifugation at 2500 × g, 10 min and resuspended in modified minimal M9 medium (Supplementary Table SIII). It was shaken at 37°C for 60 min as a recovery time for bacteria followed by addition of 250 μL of 5-fluoroindole (Santa Cruz, USA; 240 mg/mL in dimethyl sulfoxide [DMSO]). Protein production was induced with 250 μM IPTG at 30°C and harvested in 4 h. Cell pellets were resuspended in buffer A (20 mM Tris-HCl pH 7.4, 137 mM NaCl, 2.6 mM KCl, 25 mM CaCl2) supplemented with 1 mM PMSF and DNaseI (Applichem, Darmstadt, Germany). The cells were lysed by cell disruption (Branson Digital Sonifier) at 50% power 10 s on and 40 s off pulses following removal of cell debris by centrifugation (10,000 × g, 30 min, 4°C). The supernatant was loaded onto a 2 mL Pierce™ d-Gal agarose column (Thermo Fisher Scientific) that was equilibrated with 3-fold column volume of buffer A. Bound LecA was eluted with buffer B (20 mM Tris-HCl pH 7.4, 137 mM NaCl, 2.6 mM KCl, 25 mM CaCl2, 100 mM d-Gal). Protein was dialyzed in MilliQ water and TBS buffer (20 mM Tris-HCl pH 7.8, 100 mM NaCl) or MES buffer (25 mM MES pH 6, 40 mM NaCl) three times for 4 h and once overnight at 4°C, respectively. The protein solution was flash frozen and stored at −80°C.
Protein-observed fluorine (PrOF) NMR of 5FW LecA
All experiments were conducted on Bruker Ascend™700 (AvanceIII HD) spectrometer equipped with a 5 mm TCI700 CryoProbe™ in 3 mm tubes (Norell S-3-800-7) with following parameters: time domain of 1972, relaxation delay 1 s, acquisition time of 0.15 s, spectral width of 10 p.p.m. and 1024 scans resulting in measurement time of 20 minutes.
For optimization, PrOF NMR with 50, 100 or 200 μM holo 5FW LecA was recorded in TBS pH 7.8 or MES pH 6 with 10% D2O, 2 mM CaCl2 and 100 μM TFA at 285, 298 or 310 K. We considered only changes in CSP of peaks being 2-fold greater than standard deviation of fluorine resonance upon addition of 10 mM CaCl2 or 1 mM d-Gal. All data analysis, plotting and curve fitting were performed with MestReNova 11.0.0 (Mestrelab Research SL, Santiago de Compostela, Spain). All spectra were referenced and normalized to trifluoroacetic acid (TFA) as internal reference at −75.6 p.p.m. after applying the Exponential function (30 Hz) and baseline correction.
The PrOF NMR titrations of d-Gal, d-GalNAc, Ph–β-d-Gal and pNPGal were performed with 100 μM 5FW LecA in TBS pH 7.8 at 310 K. For Ca2+ titration, 5FW LecA was dialyzed against Chelex®-100 in MES pH 6 buffer at 4°C overnight.
The decreasing intensity of the unbound W42 in 5FW LecA was followed to determine Kd values of ligands. Here, we used these values to normalize the changes in W42 peak intensities (Inormalized) following the equation (1) resulting in values plotted on Y-axis.
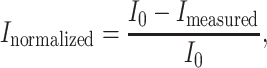 |
(1) |
where I0 was unbound W42 in the reference spectrum of protein only, Imeasured was unbound W42 peak of protein with a ligand. The Kd values were calculated according to the one-site-binding model in GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA) from three independent titrations.
Isothermal titration calorimetry
ITC was performed on a Microcal ITC200 (General Electric) at 25°C. Calcium ions were removed by extensive dialysis against 1 mM EDTA pH 7.4 (×4) followed by 150 mM NaCl (×4) and distilled water. The protein solution was lyophilized and the solid protein stored at −20°C. A solution of calcium chloride in TBS (20 mM Tris, 137 mM NaCl, 2.6 mM KCl at pH 7.4) was titrated into a calcium-free LecA solution in the same buffer. The data were analyzed according to the one-site-binding model using Microcal Origin software. Four independent titrations were performed using CaCl2 and LecA between 2 and 3 mM and 170 and 200 μM, respectively.
Competitive binding fluorescence polarization assay
The competitive binding assay was performed as reported previously (Joachim et al. 2016). In total, 10 μL of LecA (40 μM) and meta-linked fluorescein-conjugate of phenyl-galactopyranoside (20 nM) in TBS pH 7.4 supplemented with 1 mM CaCl2 (TBS/Ca2+) and 10 μL of compound-dilution series (8 mM to 62 μM, 8% DMSO) in TBS/Ca2+ buffer were mixed in a 384-well plate (Greiner Bio-One, Germany) in three technical replicates. The sealed plate was centrifuged at 300 g for 1 min and incubated at room temperature with shaking. Fluorescence intensity was measured on a PheraStar FS microplate reader (BMG Labtech GmbH, Germany; ex. 485, em. 535 nm) after 1 and 16 h. Polarization was calculated and the data were analyzed according to the four-parameter variable slope model (MARS Data Analysis Software, BMG Labtech GmbH, Germany), the top and bottom plateaus were determined from the control Me-α-d-Gal and data were reanalyzed with these values fixed.
Supplementary Material
Acknowledgments
We acknowledge Anne Imberty (CERMAV, Grenoble, France) for the plasmid pET25pa1l and fruitful discussions. We are very grateful to William C.K. Pomerantz (University of Minnesota, USA) for scientific advice with PrOF NMR.
Funding
The German Research Foundation (DFG) [Ti756/5–1, RA1944/7-1]; this was in the scope of German Research Foundation and French National Research Agency [ANR-17-CE11–0048] project ‘Glycomime’.
Conflict of interest statement
None declared.
References
- Arntson KE, Pomerantz WC. 2016. Protein-observed fluorine NMR: A bioorthogonal approach for small molecule discovery. J Med Chem. 59(11):5158–5171. [DOI] [PubMed] [Google Scholar]
- Bergmann M, Michaud G, Visini R, Jin X, Gillon E, Stocker A, Imberty A, Darbre T, Reymond JL. 2016. Multivalency effects on Pseudomonas aeruginosa biofilm inhibition and dispersal by glycopeptide dendrimers targeting lectin LecA. Org Biomol Chem. 14(1):138–148. [DOI] [PubMed] [Google Scholar]
- Cecioni S, Imberty A, Vidal S. 2015. Glycomimetics versus multivalent glycoconjugates for the design of high affinity lectin ligands. Chem Rev. 115(1):525–561. [DOI] [PubMed] [Google Scholar]
- Chemani C, Imberty A, de Bentzmann S, Pierre M, Wimmerova M, Guery BP, Faure K. 2009. Role of LecA and LecB lectins in Pseudomonas aeruginosa-induced lung injury and effect of carbohydrate ligands. Infect Immun. 77(5):2065–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioci G, Mitchell EP, Gautier C, Wimmerova M, Sudakevitz D, Perez S, Gilboa-Garber N, Imberty A. 2003. Structural basis of calcium and galactose recognition by the lectin PA-IL of Pseudomonas aeruginosa. FEBS Lett. 555(2):297–301. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Stacey RE, Dodd C, Camara M, Williams P, Winzer K. 2006. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol. 8(6):1095–1104. [DOI] [PubMed] [Google Scholar]
- Divakaran A, Kirberger SE, Pomerantz WCK. 2019. SAR by (protein-observed) 19F NMR. Acc Chem Res. 52(12):3407–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber N, Guempel U, Belz A, Gilboa-Garber N, Doyle RJ. 1992. On the specificity of the D-galactose-binding lectin (PA-I) of Pseudomonas aeruginosa and its strong binding to hydrophobic derivatives of D-galactose and thiogalactose. Biochim Biophys Acta 1116(3):331–333. [DOI] [PubMed] [Google Scholar]
- Gee CT, Arntson KE, Urick AK, Mishra NK, Hawk LM, Wisniewski AJ, Pomerantz WC. 2016. Protein-observed (19)F-NMR for fragment screening, affinity quantification and druggability assessment. Nat Protoc. 11(8):1414–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson KL, Bartlett GJ, Diehl RC, Agirre J, Gallagher T, Kiessling LL, Woolfson DN. 2015. Carbohydrate-aromatic interactions in proteins. J Am Chem Soc. 137(48):15152–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imberty A, Wimmerova M, Mitchell EP, Gilboa-Garber N. 2004. Structures of the lectins from Pseudomonas aeruginosa: Insight into the molecular basis for host glycan recognition. Microbes Infect. 6(2):221–228. [DOI] [PubMed] [Google Scholar]
- Joachim I, Rikker S, Hauck D, Ponader D, Boden S, Sommer R, Hartmann L, Titz A. 2016. Development and optimization of a competitive binding assay for the galactophilic low affinity lectin LecA from Pseudomonas aeruginosa. Org Biomol Chem. 14(33):7933–7948. [DOI] [PubMed] [Google Scholar]
- Kadam RU, Bergmann M, Hurley M, Garg D, Cacciarini M, Swiderska MA, Nativi C, Sattler M, Smyth AR, Williams P, et al. 2011. A glycopeptide dendrimer inhibitor of the galactose-specific lectin LecA and of Pseudomonas aeruginosa biofilms. Angew Chem Int Ed Engl. 50(45):10631–10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam RU, Garg D, Schwartz J, Visini R, Sattler M, Stocker A, Darbre T, Reymond JL. 2013. CH-pi "T-shape" interaction with histidine explains binding of aromatic galactosides to Pseudomonas aeruginosa lectin LecA. ACS Chem Biol. 8(9):1925–1930. [DOI] [PubMed] [Google Scholar]
- Kitevski-LeBlanc JL, Prosser RS. 2012. Current applications of 19F NMR to studies of protein structure and dynamics. Prog Nucl Magn Reson Spectrosc. 62:1–33. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. 2012. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 335(6072):1106–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck LA, Falke JJ. 1991. 19F NMR studies of the D-galactose chemosensory receptor. 2. Ca(II) binding yields a local structural change. Biochemistry. 30(17):4257–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M, Meyer B. 2001. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 123(25):6108–6117. [DOI] [PubMed] [Google Scholar]
- Renaud JP, Chung CW, Danielson UH, Egner U, Hennig M, Hubbard RE, Nar H. 2016. Biophysics in drug discovery: Impact, challenges and opportunities. Nat Rev Drug Discov. 15(10):679–698. [DOI] [PubMed] [Google Scholar]
- Rodrigue J, Ganne G, Blanchard B, Saucier C, Giguere D, Shiao TC, Varrot A, Imberty A, Roy R. 2013. Aromatic thioglycoside inhibitors against the virulence factor LecA from Pseudomonas aeruginosa. Org Biomol Chem. 11(40):6906–6918. [DOI] [PubMed] [Google Scholar]
- Sharaf NG, Gronenborn AM. 2015. (19)F-modified proteins and (19)F-containing ligands as tools in solution NMR studies of protein interactions. Methods Enzymol. 565:67–95. [DOI] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 407(6805):762–764. [DOI] [PubMed] [Google Scholar]
- Taroni C, Jones S, Thornton JM. 2000. Analysis and prediction of carbohydrate binding sites. Protein Eng. 13(2):89–98. [DOI] [PubMed] [Google Scholar]
- Tobola F, Lelimousin M, Varrot A, Gillon E, Darnhofer B, Blixt O, Birner-Gruenberger R, Imberty A, Wiltschi B. 2018. Effect of noncanonical amino acids on protein-carbohydrate interactions: Structure, dynamics, and carbohydrate affinity of a Lectin engineered with fluorinated tryptophan analogs. ACS Chem Biol. 13(8):2211–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Hauck D, Hoffmann M, Sommer R, Joachim I, Müller R, Imberty A, Varrot A, Titz A. 2017. Covalent Lectin inhibition and application in bacterial biofilm imaging. Angew Chem Int Ed Engl. 56(52):16559–16564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Sommer R, Hinsberger S, Lu C, Hartmann RW, Empting M, Titz A. 2016. Novel strategies for the treatment of Pseudomonas aeruginosa infections. J Med Chem. 59(13):5929–5969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


