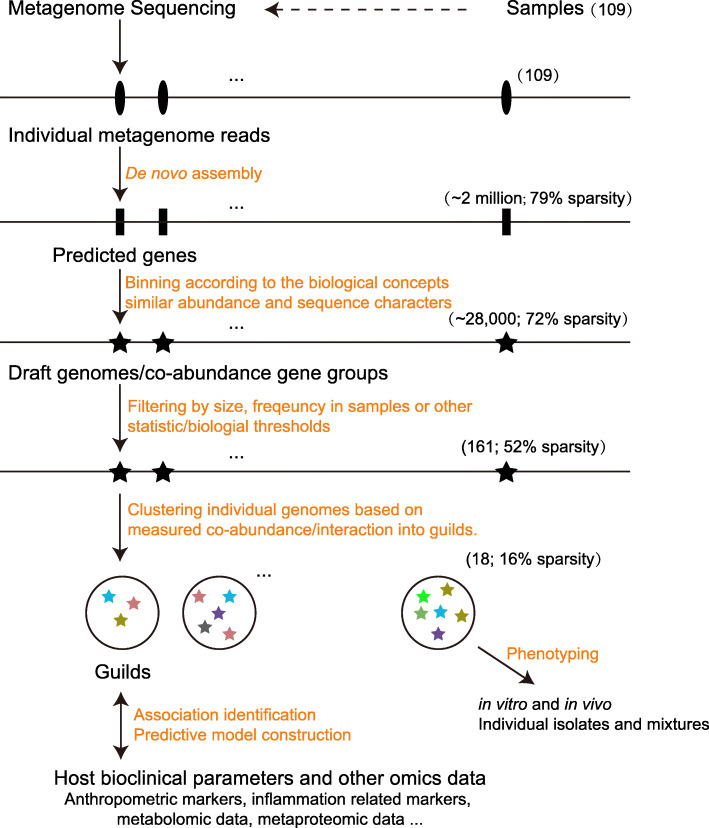
Fig. 1.
The reduction of dimensionality and sparsity from raw metagenomic dataset to genes, genomes, and guilds. In our PWS example, ~ 2 million non-redundant microbial genes were predicted from the 109 metagenomes. Seventy-nine percent of values in the corresponding abundance matrix of these genes were zeros. These non-redundant microbial genes were further binned into ~ 28,000 draft genomes based on their abundance correlations across the 109 samples. In the corresponding abundance matrix of these draft genomes, 72% of values were zeros. We then selected 161 prevalent bacterial genomes, each with more than 700 bacterial genes and shared by more than 20% of the samples. In the corresponding abundance matrix of these 161 genomes, 52% of values were zeros. Eighteen guilds were identified by clustering these prevalent bacterial genomes. In the corresponding abundance matrix of these 18 guilds, 16% values were zeros