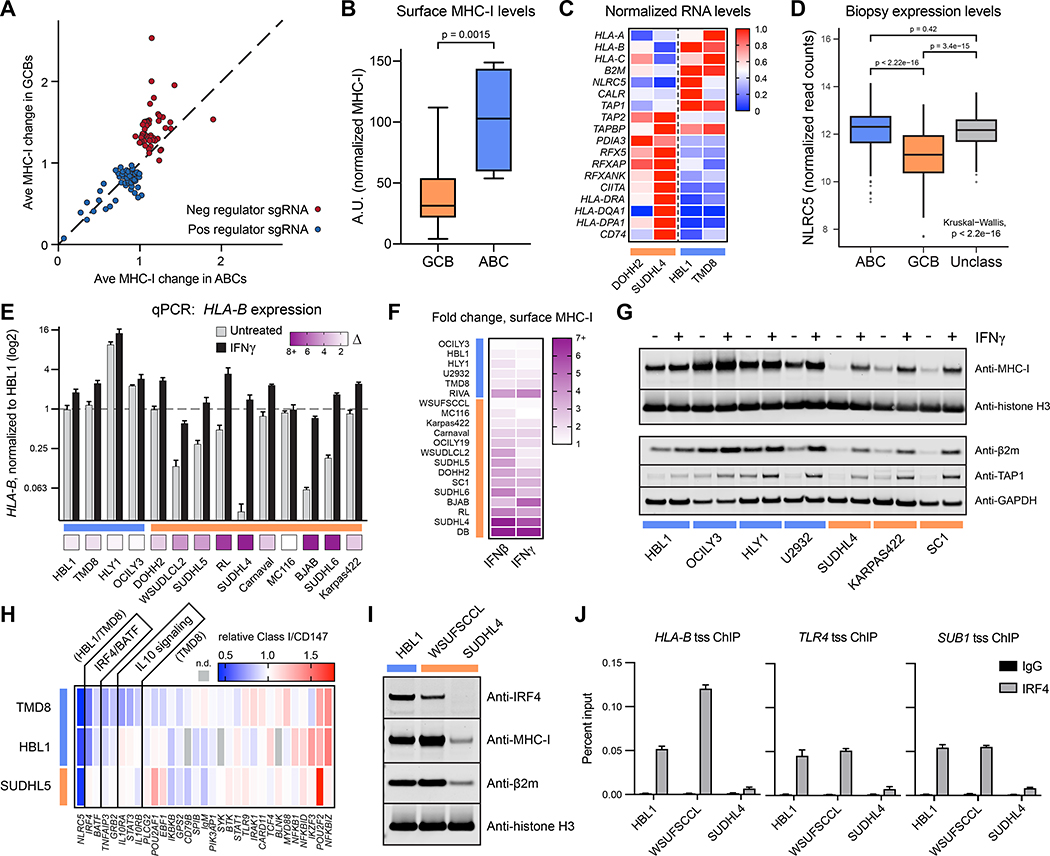
Figure 5. Activated B cell-like DLBCLs drive MHC-I antigen presentation.
(A) The fold change in surface MHC-I of a gene ablation is plotted as an average between GCBs (SUDHL5, BJAB) and between ABCs (HBL1, TMD8). The diagonal line indicates an equivalent response between the subtypes. (B) A panel of 20 DLBCL cell lines were measured for surface MHC-I by flow cytometry, and MFI were normalized to fluorescent beads. For entire figure: GCBs orange, ABCs blue. (C) mRNA was isolated from the indicated cells and subjected to RNAseq. Each transcript is normalized to the highest FPKM value of the four lines. (D) RNAseq was performed on DLBCL patient tumor biopsies, and NLRC5 gene expression is displayed as log transformed normalized read counts (n = 574 patients). (E) Cells were treated with 0 or 500U/mL IFNγ for 48 hours, at which point RNA was harvested for qPCR of HLA-B transcripts. Bottom boxes represent fold increase. (F) Cells were treated with 0 or 500U/mL of IFNβ or IFNγ and stained for surface MHC-I by flow cytometry after 48 hours. (G) Same as E, but whole cell lysates were extracted and analyzed by immunoblotting. (H) TMD8, HBL1, and SUDHL5 cells were transduced with sgRNA for the indicated genes, and surface MHC-I complexes were measured compared to non-targeting controls while cells retained viability. (I) DLBCL whole cell lysates were blotted with antibodies to the indicated proteins. (J) HLA-B, TLR4, and SUB1 transcriptional start site (tss) occupancy by IRF4 was examined by ChIP. For entire figure, means with standard deviations are plotted; minimum n = 3. See also Figure S3.