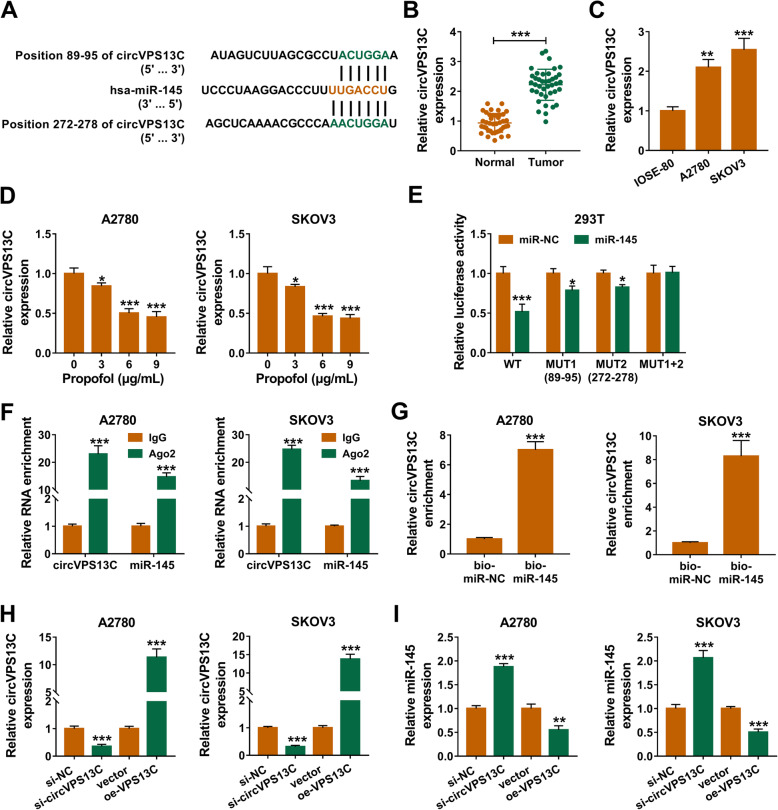
Fig. 4.
CircVPS13C directly interacts with miR-145. a There existed two sites in circVPS13C that were complementary with miR-145 (predicted by circinteractome database), including position 89–95 and position 272–278. b and c The abundance of circVPS13C in adjacent normal tissues, ovarian cancer tissues, IOSE-80 cell line and two ovarian cancer cell lines was detected by qRT-PCR. d Ovarian cancer cells were exposed to different concentrations of Propofol, and the level of circVPS13C was detected by qRT-PCR. e The wild-type sequence of circVPS13C that contains the two complementary sites with miR-145, the single mutant sequence (89–95) of circVPS13C, the single mutant sequence (272–278) of circVPS13C and the double mutant sequence of circVPS13C were amplified and cloned into luciferase reporter vectors, generating WT, MUT1 (89–95), MUT2 (272–278) and MUT1 + 2, respectively. Dual-luciferase reporter assay was used to test which position in circVPS13C could directly bind to miR-145, and the luciferase activity was detected in 293 T cells co-transfected with these reporter plasmids and miR-NC or miR-145. f RIP assay was used to test whether there existed spatial interaction between miR-145 and circVPS13C in ovarian cancer cells. g The interaction between miR-145 and circVPS13C was tested by RNA-pull down assay. h and i Ovarian cancer cells were transfected with si-NC, si-circVPS13C, vector or oe-circVPS13C. The levels of circVPS13C and miR-145 in transfected ovarian cancer cells were examined by qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001