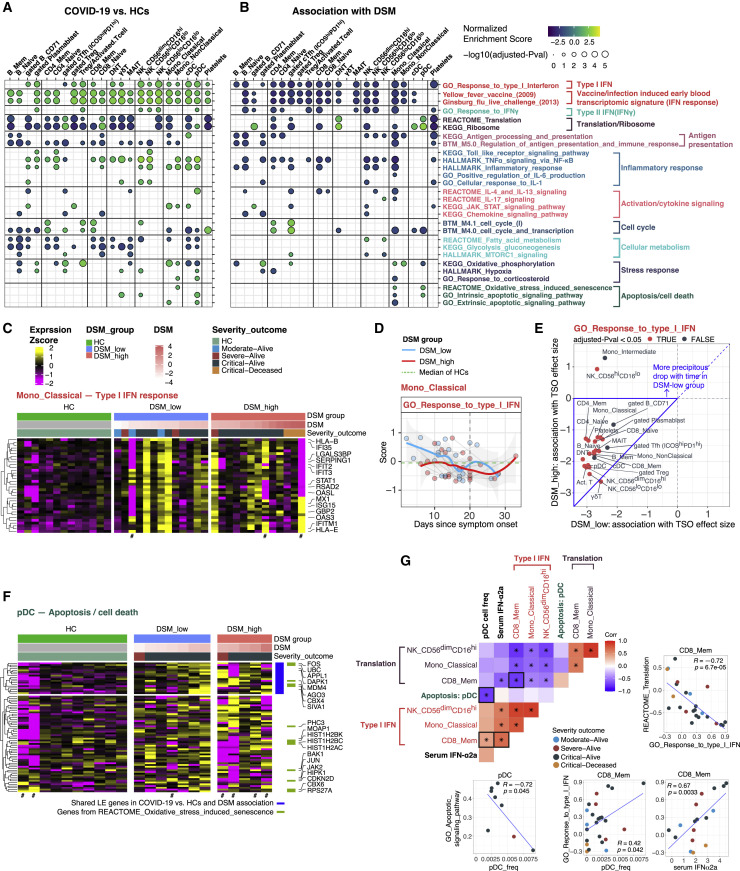
Figure 3.
Cell-type-specific gene expression signatures of COVID-19 disease status and severity
(A) Gene set enrichment analysis (GSEA) result of COVID-19 versus HCs at T0. Selected gene sets (rows, see STAR methods) are grouped into functional/pathway categories. Differential expression moderated t statistics were computed from a linear model accounting for age, and experimental batch (TSO not accounted for as TSO does not exist for healthy individuals). Dot color denotes normalized gene set enrichment score (NES) and size denotes -log10(adjusted p value). p values were adjusted using the Benjamini-Hochberg method. The sign of the NES corresponds to increase/decrease of change in COVID-19 compared to HCs. Cell populations (columns) are grouped by lineage/type. See Table S4A for detailed results of all gene sets which passed the adjusted p value cutoff of 0.2.
(B) Similar to (A) but the enrichment analysis was performed based on association with DSM at T0, and the model controlled for TSO. The sign of the NES score denotes the sign of the association with DSM. See also Table S4B.
(C) Heatmap of type I IFN response in classical monocytes. Heatmap showing the scaled average mRNA expression (within the indicated cell cluster/population for a given subject) of shared leading-edge (LE) genes from the GSEA analysis of COVID-19 versus HCs (see A) and association with DSM (see B). Selected top LE genes are labeled by gene symbol and only the sample from the first time point for each patient (T0) is shown. For gene expression heatmaps, subjects are grouped by HC and DSM classes; also shown are the DSM value and severity-outcome classification of each patient (top of the heatmaps). Subjects with a small number of cells not included in the GSEA test are marked by a # (cell number <8).
(D) Per-sample gene set signature scores of the GO_Response to type I IFN gene set versus TSO (in days) in DSM-low and DS-high groups. Classical monocyte is shown as an example. Individual samples are shown as dots and longitudinal samples from the same individual are connected by gray lines. Trajectories (using LOESS smoothing, see STAR methods) were fitted to the groups separately with the shaded areas representing standard error. The median score of age- and gender-matched HCs is shown as a green dotted line. Gene set scores were calculated using the union of LE genes from both the timing (TSO) and disease severity (DSM) association models using gene set variation analysis (see STAR methods).
(E) Scatterplot comparing the effect size of association between TSO and the GO_Response to type I IFN gene set in the DSM-high (y axis) and DSM-low (x axis) patients. Each dot corresponds to a cell type/cluster. The effect size reflects the change of type I IFN signature enrichment with time (effect size <0 corresponds to the enrichment decreasing with time). Cell types are colored by their statistical significance (adjusted p value <0.05) of whether the association with time is significantly different between the two DSM groups (based on a model with a DSM-TSO interaction term, see STAR methods). The area framed by blue borders corresponds to cell types with more precipitous drop of IFN signature over time in the DSM-low than DSM-high groups.
(F) Heatmap of apoptosis/cell death signature in pDCs. Similar to (C), but instead of the shared LE genes, all LE genes from COVID-19 versus HCs and association with DSM are shown. Shared LE genes and genes in the REACTOME_Oxidative stress-induced senescence gene set are annotated on the right.
(G) Heatmap showing the sample-level pairwise Pearson correlations among serum IFN-α2a level, pDC frequency, the apoptosis signature score in pDCs, as well as the IFN-I and protein translation signature scores in classical monocyte, CD56dimCD16hi NK, and CD8 memory T cell clusters (∗p value <0.05). Selected scatterplots are shown, and the corresponding entry in the heatmap is indicated using bold borders. Samples were filtered to remove those with fewer than 8 cells or less than 40,000 unique molecular identifiers (UMI) in the pseudobulk library. Note that not all subjects had IFN-α2a measurements. Each dot indicates a subject and is colored by the severity-outcome category.