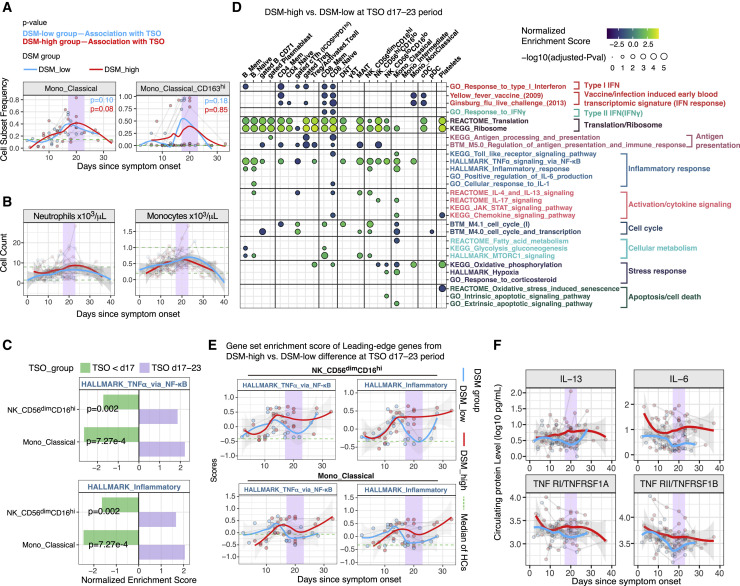
Figure 5.
Analyses of timing effects suggest a late immune response juncture
(A) Time course of monocyte subset frequencies in DSM-low and DSM-high groups. Classical monocyte is expressed as fraction of total PBMCs; the CD163hi classical monocyte subset is expressed as a fraction of classical monocytes. The median cell frequency of age- and gender-matched HCs is shown as a green dotted line. Individual samples are shown as dots and longitudinal samples from the same individual are connected by gray lines. P values shown are from mixed effect linear models indicating the statistical significance of the timing effect (i.e., TSO) accounting for age and experimental batch in DSM-low and DSM-high groups, respectively. Trajectories (using LOESS smoothing) were fitted to DSM-low and DSM-high groups separately. TSO = days 17–23 period is highlighted with purple.
(B) Similar to (A) but showing the absolute blood neutrophil and monocyte counts. The two green dotted lines mark the approximate reference range of cell counts in healthy adults. The shaded areas around trajectories denote standard error.
(C) Effect size (normalized enrichment score from GSEA) comparison of the period before day 17 (TSO <day 17, green) and during the TSO = days 17–23 period (purple) for inflammatory related gene sets. Effect sizes correspond to differences between DSM-high versus DSM-low groups (e.g., effect size <0 corresponds to the gene signature being less enriched in the DSM-high group than DSM-low group). p values shown are FDR adjusted (via the Benjamini-Hochberg method) from the test reflecting the temporal changes in the difference between DSM-high and DSM-low groups from before the day 17 period to the days 17–23 time window (see STAR methods).
(D) Similar to Figure 3A, but showing GSEA results for assessing differences between the DSM-high versus DSM-low groups using only samples from days 17–23 since symptom onset. See Table S4E for detailed results of these selected gene sets and all sets that passed the adjusted p value cutoff of 0.2.
(E) Time course of gene set signature scores of inflammatory related gene sets in DSM-low and DSM-high patient groups in CD56dimCD16hi NK cells and classical monocytes. The gene set signature scores were calculated using the LE genes identified from the enrichment analysis shown in (C) above to highlight differences between the DSM-high versus DSM-low groups during the days 17–23 period. The median score of age- and gender-matched HCs is shown as a green dotted line. The shaded areas around trajectories denote standard error.
(F) Time course of serum protein levels from DSM-low and DSM-high patients, respectively. Similar to (E). Top 4 serum proteins significantly different between DSM-high versus DSM-low groups during the days 17–23 period are shown. See Table S6 for the full list of differentially expressed proteins.