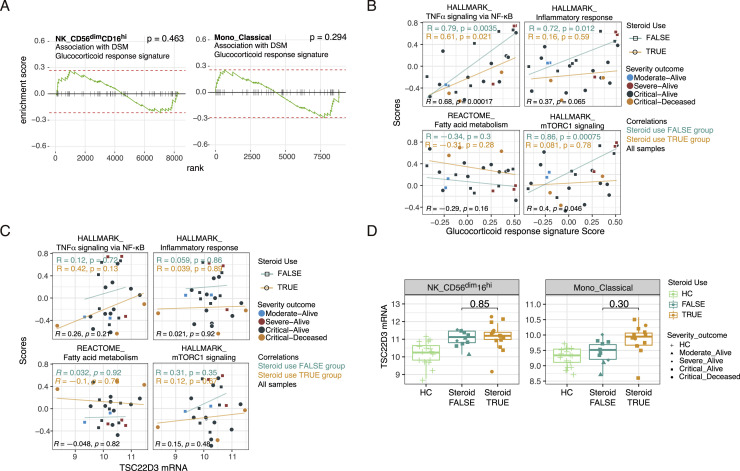
Figure S5.
Exogeneous corticosteroid treatment is not a major driver of cell-type-specific gene expression signatures associated with disease severity, related to Figure 4
(A) GSEA results for glucocorticoid response signature (see STAR methods on how the signature was derived) in DSM association model. Positive enrichment corresponds to higher level in the DSM-high samples. (B and C) Scatterplot showing the correlations between the indicated signature scores (computed using GSVA) and the glucocorticoid response signature score (B) or the TSC22D3 mRNA expression level (C) in CD56dimCD16hi NK cells. Pearson correlations and p values are reported for patients with corticosteroid use (orange; steroid use TRUE) and those without (cyan; steroid use FALSE) as well as for all samples (black). Each dot indicates a subject, shaped by steroid use status and colored by the severity-outcome category. (D) TSC22D3 mRNA expression levels of CD56dimCD16hi NK cells and classical monocytes in HC, no steroid-use and steroid-use COVID-19 patient groups. P values from testing for differences between the no steroid-use and steroid-use groups are shown. P values were obtained from an ANOVA test accounting for DSM, TSO, age and experimental batch.