Figure 2. Distribution of species with proteomic data sets (P), genome assemblies (G), and genome annotations (A) across phylogenetic clades.
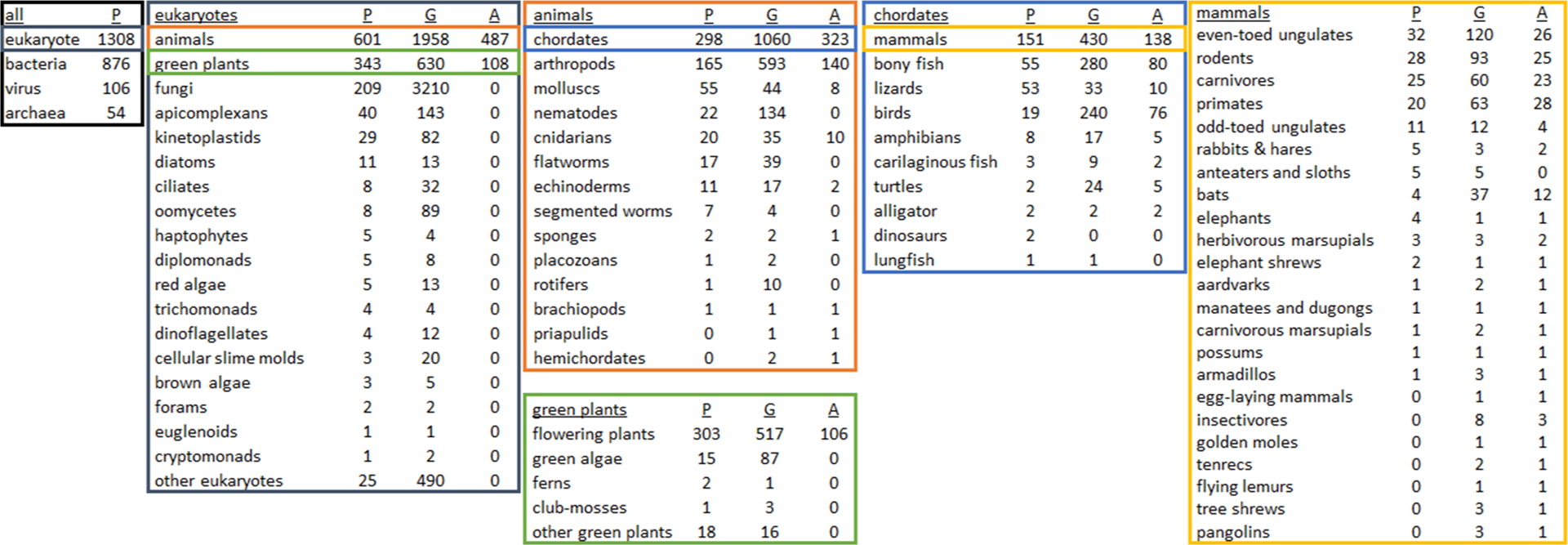
This series of charts was created by cross-referencing taxon IDs of genome assemblies on Genbank and genome annotations on RefSeq (valid as of 12 March 2020) with published proteomic data sets on the Proteomics Identifications Database (PRIDE; as of 17 January 2020) in order to emphasize clades lacking proteomic analysis, or clades that should be focused on for genomic sequencing and annotation. Specific to Eukaryotes, there are approximately 1308 species with published proteomic results, 6718 with available genomes and 595 species with complete genomic annotations. Note that these charts do not show the total number of species per clade (e.g., 950 000 named insect species65), or the number of proteomic data sets per species (e.g., 68 % of the 12 660 PRIDE data sets are human and mouse). For this cataloging, we focused only on genome and annotation resources at NCBI and acknowledge that for non-animal non-flowering plants, there are other more appropriate repositories such as the Joint Genome Institute. Terms in tables are accepted common or scientific NCBI designations, except in a few cases under ‘mammals’ (the full mammal table with taxon IDs is available in Supplemental Table S1), and that these numbers may not be exact due to self-reporting on PRIDE and matching taxon identifiers between sources (note that the two dinosaur proteomic data sets are from Brachylophosaurus canadensis and Tyrannosaurus rex).
