Figure 3.
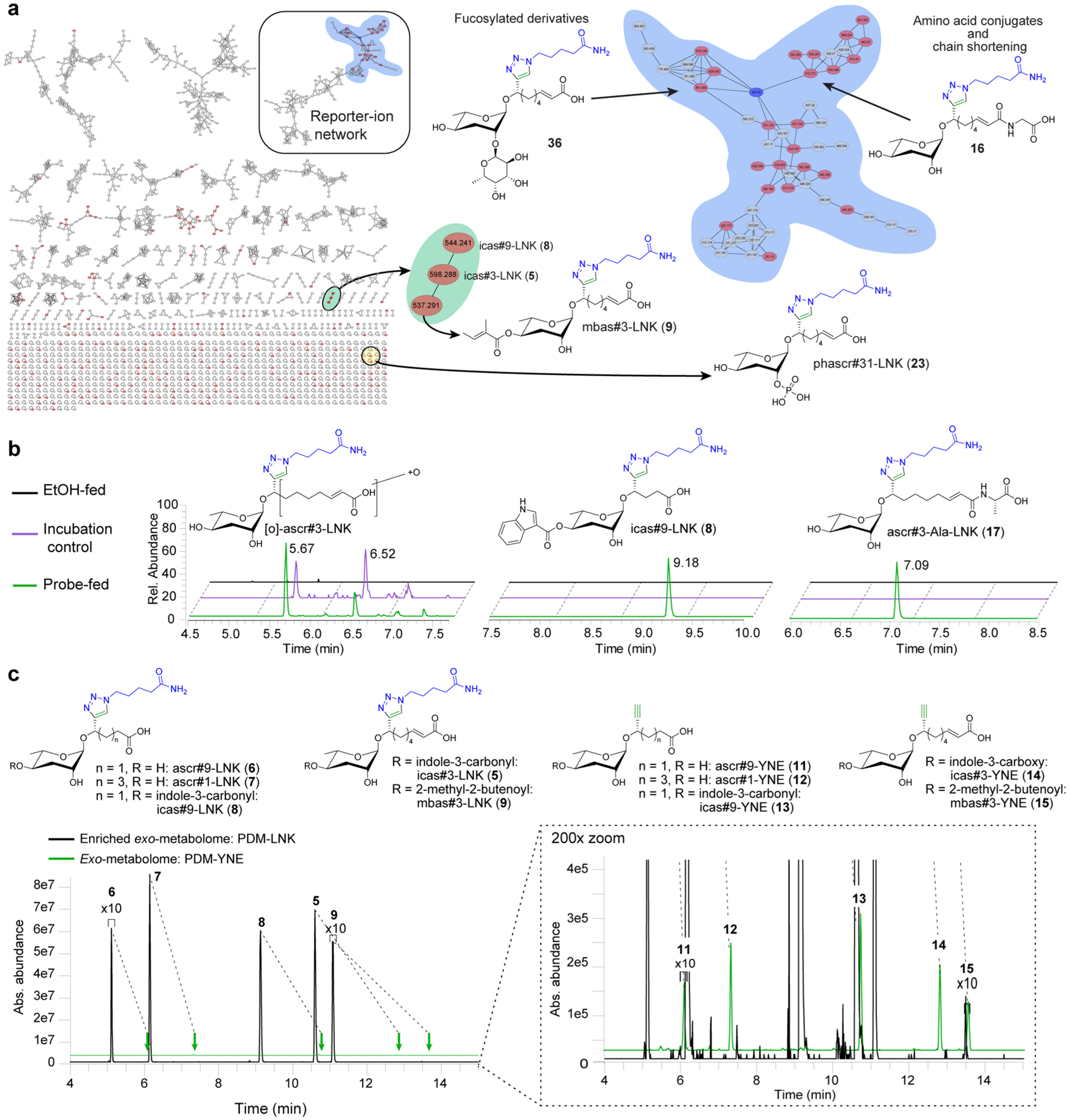
DIMEN of ascr#3-YNE-fed C. elegans. (a) Molecular network resulting from DIMEN analysis of ascr#3-YNE-treated C. elegans cultures. The blue node is ascr#3-LNK and the red nodes are differential between probe-fed and incubation control data sets. These include analogues of known ascarosides such as mbas#3-LNK (9) as well as several new compound families, e.g., a set of fucosylated PDMs, amino-acid ligated ascarosides, and phosphorylated ascarosides. (b) Example EICs for probe-derived artifacts detected in both the probe-fed and incubation control data sets that originate from abiological transformations (e.g., [O]-ascr#3-LNK, a product of nonenzymatic oxidation of ascr#3), PDMs only present in probe-fed data sets representing known ascaroside derivatives (e.g., icas#9-LNK (8), a derivative of the known icas#9), and PDMs only present in probe-fed data sets that represent novel metabolites (e.g., ascr#3-Ala-LNK (17)). (c) EICs filtered for alkyne-functionalized PDMs (PDM-YNE) in exo-metabolome (green), and linker-bound PDMs (PDM-LNK) in enriched exo-metabolome (black). DIMEN improves signal intensity >200-fold between pairs of PDM-YNE and PDM-LNK, due to enrichment and improvement of ionization efficiency (Figure S4d).
