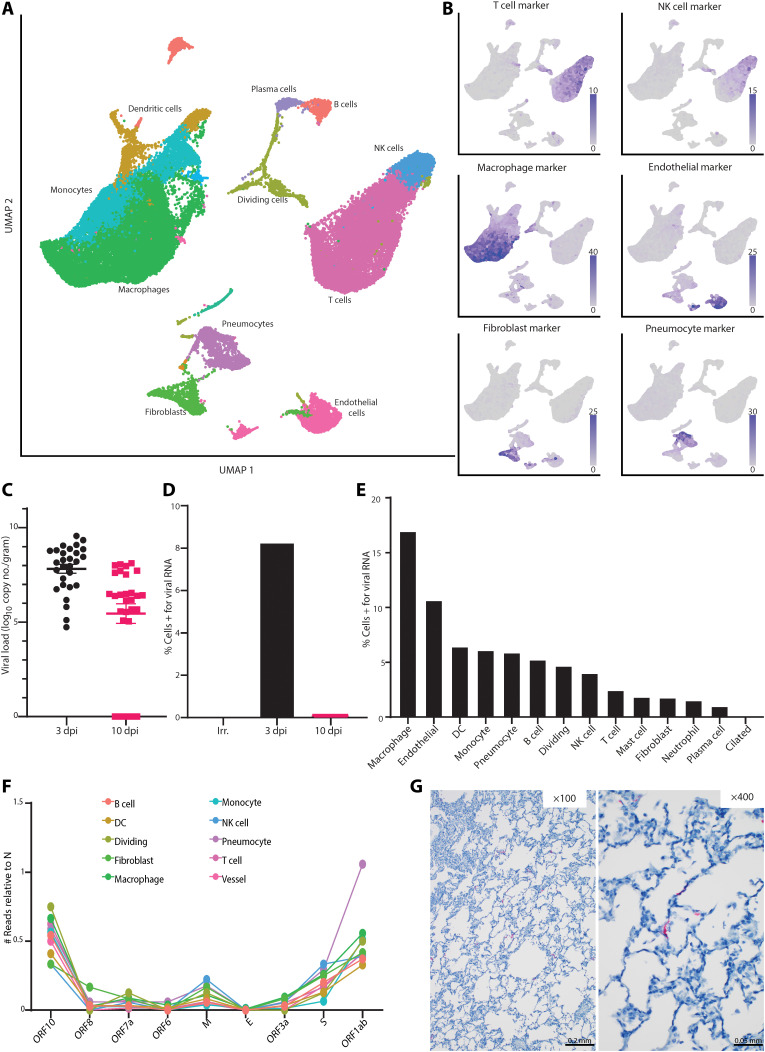
Fig. 3. Single-cell sequencing reveals viral dynamics in lung tissue.
(A) UMAP projection of scRNA-seq data from whole lung sections from all 10 animals combined. Each point is an individual cell; colors are based on cell type annotation. Cell names are shown next to their largest cluster. NK, natural killer. (B) Validation of cell type identities using marker gene sets. The intensity of the purple color represents higher expression of the indicated marker set. Gray coloring indicates that the cell did not express any genes in the marker set. (C) Viral load in cells isolated from the lungs was evaluated via qRT-PCR for gRNA and grouped for all lobes across each animal in the indicated groups. (D) The percentage of cells identified by scRNA-seq that were positive for any reads aligning to the viral genome by dpi is reported. (E) Percentage of cells from the 3-dpi samples positive for any reads aligning to the viral genome grouped by cell type. DC, dendritic cell. (F) The number of cells grouped by cell type with reads aligning to other locations across the viral genome, all normalized to the number of cells expressing nucleocapsid (N) gene. M, membrane; E, envelope; S, spike. Genes are ordered from the 3′ to 5′ end of the SARS-CoV-2 genome. (G) ISH for viral spike RNA in lung tissues at 3 dpi. Viral RNA staining is shown in red at ×100 magnification (scale bar: 0.2 mm) and ×400 magnification (scale bar: 0.05 mm).