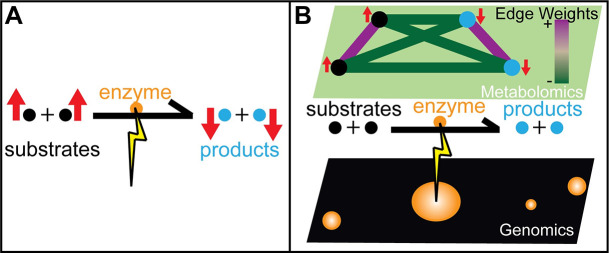
Fig 2. Latent covariance structure in the metabolome is induced by genetic perturbations of an enzyme.
(A) Pathway visualization can unveil bottlenecks in metabolite flow when enzymes fail to function due to biallelic pathogenic mutations. Substrates are perturbed upwards and products are perturbed downwards around an affected enzyme. (B) Gaussian Markov Random Field (GMRF) networks model covariation relationships between metabolite perturbations, where each edge indicates a non-zero partial correlation between two given metabolites. When an enzyme’s function is deficient, substrate levels are perturbed upwards, product levels are perturbed downwards. This latent covariance structure within the GMRF model is characterized by positive covariance edges between substrates and between products and negative covariance edges between substrates and products.