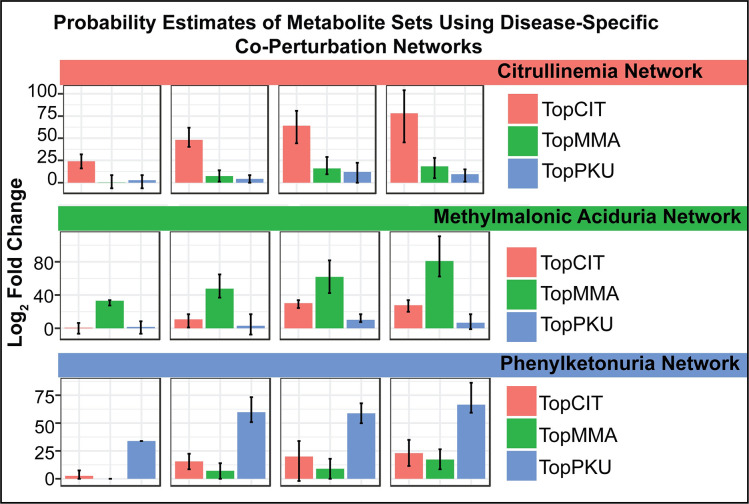
Fig 7. The probability of a metabolite set depends on the disease-specific network used in the encoding process.
The probability of the 5, 10, 15 or 20 most perturbed metabolites across (A) citrullinemia samples (B) methymalonic aciduria samples or (C) phenylketonuria samples is much larger when using the network learned from patients with citrullinemia, methylmalonic aciduria, and phenylketonuria, respectively, compared to the probability of the same metabolite set when using a permuted network. Error bars indicate signal observed across several network folds when using a leave-one-out cross validation scheme for network learning and scoring.