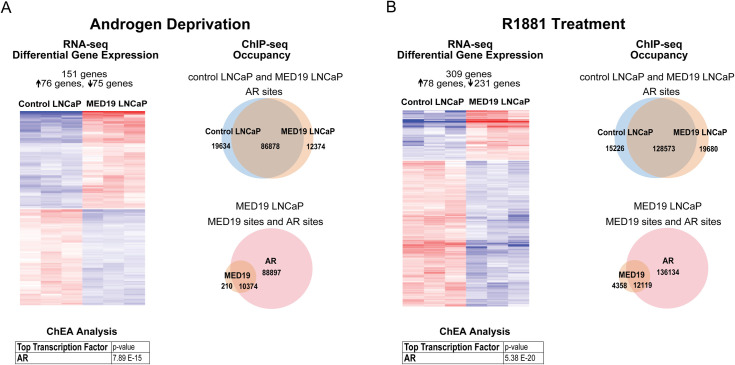
Fig 4. MED19 overexpression causes a selective shift in gene expression and in the AR cistrome under androgen deprivation and with R1881 treatment.
MED19 LNCaP cells and control LNCaP cells were cultured under androgen deprivation for 3 days, and cells were treated with ethanol vehicle or R1881 (10 nM for 16 h for RNA-seq and 100 nM for 4 h for ChIP-seq). RNA-seq with ribodepletion was performed in biological triplicate. ChIP-seq for FLAG-MED19, AR, and H3K27ac was performed in biological triplicate, with the exception of ChIP-seq for AR in control LNCaP cells + R1881, where one sample was excluded from the analyses because of low signal. A) (Left) Heatmap of differentially expressed genes with MED19 overexpression (fold change ≥1.5, p-adj ≤0.05) for androgen deprivation, associated with AR as the top regulatory transcription factor from ChEA. (Right) Number and overlap of occupancy sites under androgen deprivation for AR in control LNCaP cells and MED19 LNCaP cells (top) and for AR and MED19 in MED19 LNCaP cells (bottom). B) (Left) Heatmap of differentially expressed genes with MED19 overexpression (fold change ≥1.5, p-adj ≤0.05) for R1881 treatment, associated with AR as the top regulatory transcription factor from ChEA. (Right) Number and overlap of occupancy sites with R1881 treatment for AR in control LNCaP cells and MED19 LNCaP cells (top) and for AR and MED19 in MED19 LNCaP cells (bottom).