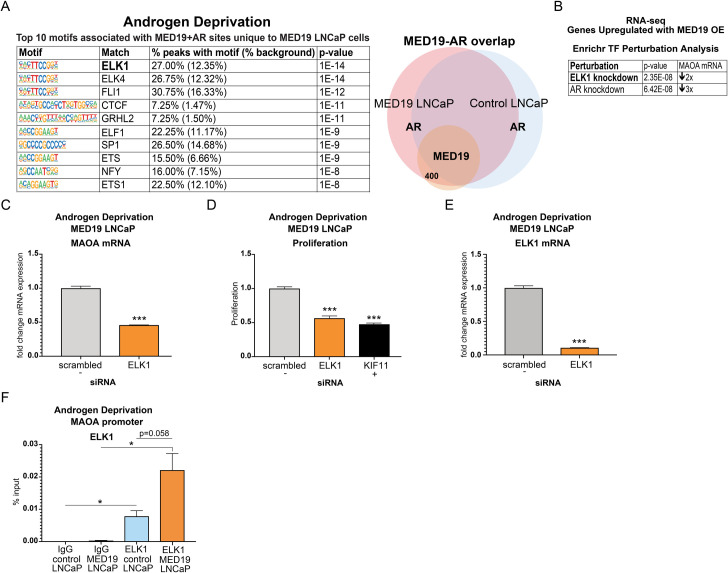
Fig 8. ELK1 is enriched at sites of AR and MED19 occupancy unique to MED19 LNCaP cells under androgen deprivation, occupies the MAOA promoter, and controls MAOA expression and androgen-independent growth.
MED19 LNCaP cells and control LNCaP cells were cultured under androgen deprivation for 3 days and treated with ethanol vehicle. RNA-seq with ribodepletion was performed in biological triplicate. ChIP-seq for FLAG-MED19, AR, and H3K27ac was performed in biological triplicate, with the exception of ChIP-seq for AR in control LNCaP cells + R1881, where one sample was excluded from the analyses because of low signal. A) The top 10 enriched transcription factor motifs under androgen deprivation associated with sites of AR and MED19 occupancy in MED19 LNCaP cells where AR is present only in MED19 LNCaP cells are shown, with ELK1 as the top associated transcription factor (labeled diagram of occupancy sites, right). B) ELK1 knockdown is the top hit from Transcription Factor Perturbation from Enrichr associated with genes upregulated by MED19 overexpression under androgen deprivation from the RNA-seq study; AR knockdown is also strongly associated. Fold downregulation of MAOA mRNA with associated ELK1 knockdown (GSE 34589) or associated AR knockdown (GSE 22483) is shown. C) ELK1 was depleted by siRNA and mRNA expression of MAOA in MED19 LNCaP cells cultured in androgen-depleted media was measured after 5 days (fold change expression normalized to RPL19, with MAOA mRNA expression with scrambled siRNA treatment set as “1”). Experiment was performed in biological triplicate, with representative results shown. D) ELK1 was depleted by siRNA and proliferation of MED19 LNCaP cells in androgen-depleted media was evaluated after 7 days, normalized to proliferation with scrambled siRNA (negative control, light grey). KIF11 knockdown is included as a positive control (black). Experiment was performed in biological duplicate, with representative results shown. E) Validation of ELK1 knockdown, with ELK1 mRNA measured as in C (with ELK1 mRNA expression with scrambled siRNA treatment set as “1”). F) ChIP-qPCR for ELK1 at the MAOA promoter overlapping with published ARE. *p < 0.05; **p < 0.01; and ***p < 0.001.