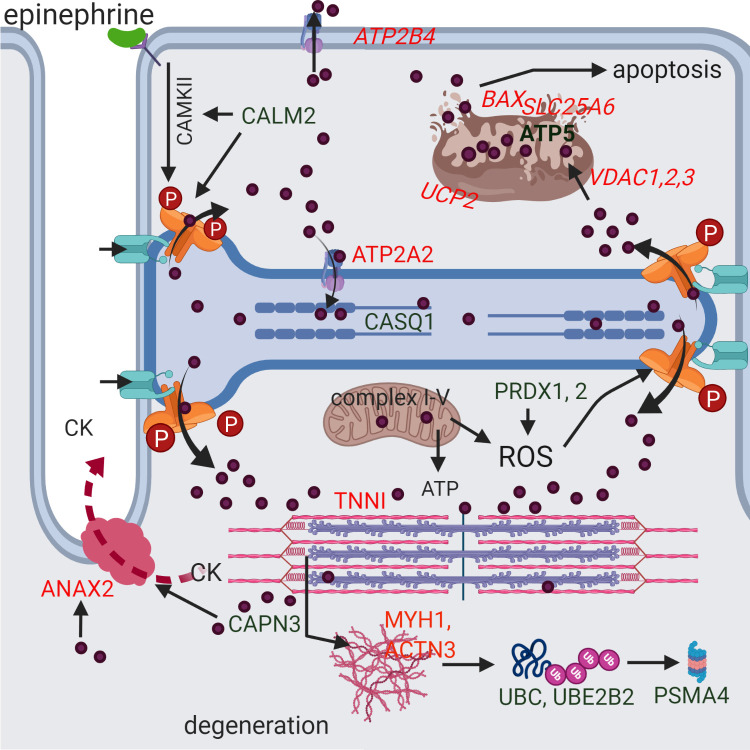
Fig 2. A potential scenario for the development of rhabdomyolysis in RER-susceptible horses derived from differentially expressed proteins and genes.
Green indicates increased and red decreased differential expression. Genes are italicized. A stressful environment is proposed to induce hyper-phosphorylation of RYR1 through beta adrenergic (epinephrine mediated) activation of calmodulin kinase II (CAMKII) which is stimulated by the DEP calmodulin (CALM2). This and other potential post-translational modifications of RYR1 (oxidation/nitrosylation) allow excessive Ca2+ (purple circles) release of high sarcoplasmic reticulum Ca2+ stores in RER-susceptible horses through RYR1 (orange) when RYR1 opening is stimulated during exercise by the voltage gaited Ca2+ channel in the t-tubule (turquoise). Myoplasmic Ca2+ interacts with troponin I (TNNI) and, when not adequately pumped back into the sarcoplasmic reticulum (ATP2A2) or out of the cell (ATP2B4), Ca2+ produces persistent contracture of the sarcomere (pink and purple). Mitochondria buffer myoplasmic Ca2+ through VDAC uptake where Ca2+ stimulates ATP production. However, in excess, Ca2+ uncouples (UCP2) oxygen consumption from electron transport and ATP production and releases reactive oxygen species (ROS). Antioxidants such as peroxiredoxin (PRDX1, PRDX2) counteract ROS to prevent oxidative stress. Excessive mitochondrial matrix Ca2+ results in formation of the membrane transition pore (BAX, SLC25A6, ATP5), loss of membrane potential, release of cytochrome C and apoptosis. Calcium activation of proteases such as calpain (CAPN3) results in degradation of myofilaments, cellular proteins and cell membranes resulting in the release of muscle proteins such as creatine kinase (CK) into the blood stream. Calcium activated ANAX2 participates in repair of membranes that have increased dystrophin (DMD) and syntropin (SNTB1) content. Degraded proteins are ubiquitinated (UBC, UBE2B2) and processed in the proteasome (PSMA4).