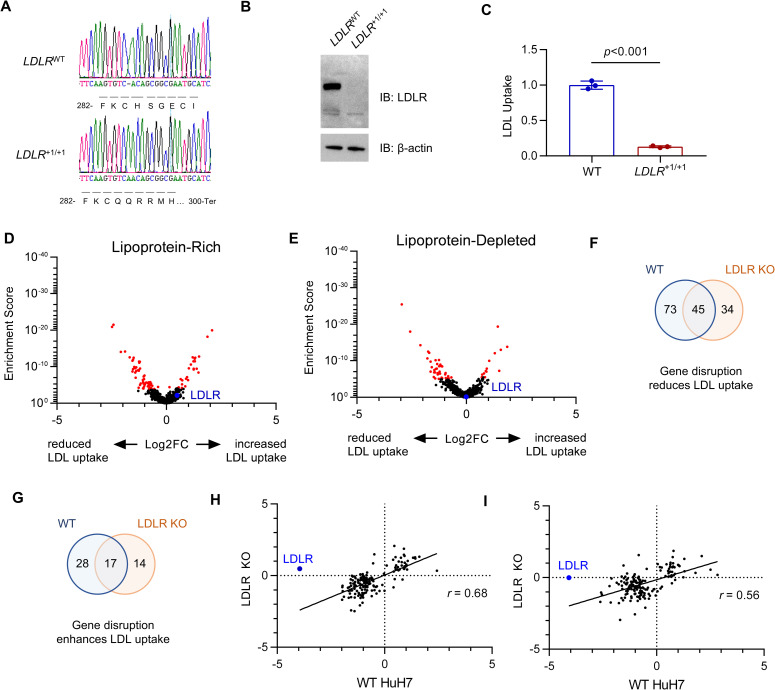
Fig 4. Orthogonal CRISPR screen for modifiers of LDL uptake by LDLR-deleted HuH7 cells.
(A) Genotyping at the genomic DNA target site, (B) immunoblotting, and (C) quantification of LDL uptake by flow cytometry for a single cell HuH7 clone targeted by CRISPR at the LDLR locus. (D-E) Volcano plots displaying MAGeCK gene level enrichment scores and log2 fold change for each gene tested in the secondary gRNA library, under lipoprotein-rich (D) or lipoprotein-depleted (E) cultured conditions, with genes identified with FDR<5% displayed in red. (F-G) Venn diagrams demonstrating the overlap in genes identified from HuH7 WT and LDLR KO cells for genes whose disruption was associated with reduced (F) or enhanced (G) LDL uptake. (H-I) Comparison of effect size on LDL uptake in WT and LDLR KO cells under lipoprotein-rich (H) or lipoprotein-depleted (I) conditions for each gene showing a significant effect in either background.